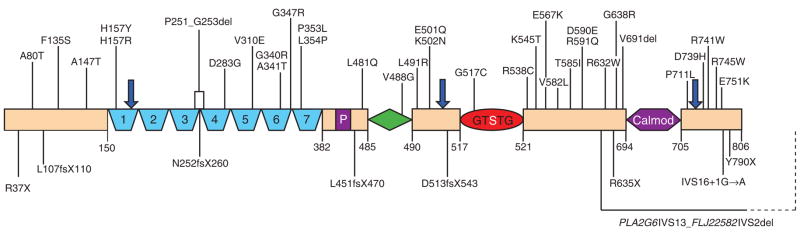
Figure 1.
Schematic showing iPLA2-VIA protein structure7 and mutation-derived changes. Variant 1 is shown with seven ankyrin repeats (blue), a proline-rich motif (purple box), a glycine-rich nucleotide binding motif (green diamond), a lipase motif (red with the active site, Ser519, highlighted), a proposed C-terminal Ca2+-dependent calmodulin binding domain (purple) and three putative caspase cleavage sites (blue arrows). Numbers indicate amino acids. Substitutions or deletions of amino acids not altering the reading frame are shown above the diagram; nonsense mutations, splice mutations and deletions causing frameshifts are shown below. A deletion encompassing exons 14 through 17, the 3′of PLA2G6 and exons 1 and 2 of a downstream putative transcript, FLJ22582, is shown at the C terminus.