Figure 1.
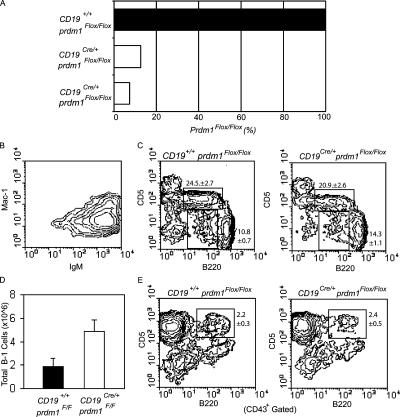
Prdm1 gene deletion and phenotypic analysis of PerC and splenic B-1 cells. (A) Quantitative real-time PCR for prdm1 performed on genomic DNA from two purified CD19Cre/+prdm1Flox/Flox (open bars) and one littermate control (filled bar) PerC B-1 cell cultures. Primers were designed to amplify the floxed, but not the deleted, allele (see Materials and methods), and DNA loading was normalized to the peptidyl prolyl isomerase A gene. Percentages of prdm1Flox/Flox amplified DNA are shown. (B) Representative flow cytometry analysis from one purified CD19Cre/+ prdm1Flox/Flox B-1 cell culture used for deletion analysis in A, stained with antibodies against IgM and Mac-1. (C) Total PerC cells from CD19Cre/+ prdm1Flox/Flox mice (right) and littermate control (left) mice stained with antibodies against B220 and CD5. Upper gate is B-1a, and the lower gate is B-1b. Mean ± SEM, n = 7. (D) Bar graph shows averages and SEM of total PerC B-1 cells (IgM+Mac-1+), n = 7. (E) Total splenocytes stained with antibodies for CD43, B220, and CD5. B220 and CD5 expression on CD43+ cells are shown. Mean ± SEM, n = 4.