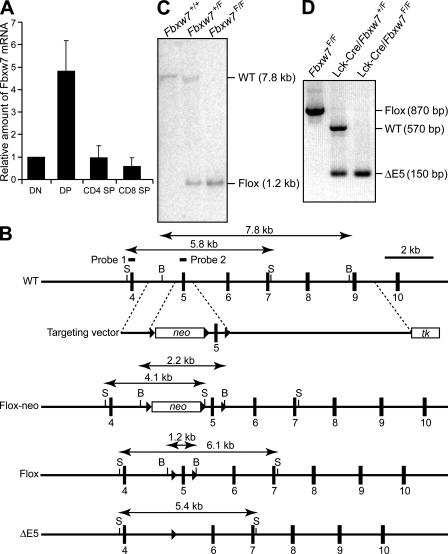
Figure 1.
Generation of mice with T cell–specific deficiency of Fbxw7. (A) RT and real-time PCR analysis of the relative abundance of Fbxw7 mRNA in DN, DP, CD4 SP, and CD8 SP mouse thymocytes isolated by flow cytometry. Data are means ± SD of values from three independent experiments. (B) Schematic representations of the wild-type mouse Fbxw7 allele, the targeting vector, the Fbxw7 allele with a loxP-neo cassette (Flox-neo), the floxed Fbxw7 allele (Flox), and the floxed Fbxw7 allele after removal of exon 5 by Cre recombinase (ΔE5). The expected sizes of bands in a Southern blot analysis with probe 1 (for StuI [S] fragments) or probe 2 (for BamHI [B] fragments) are indicated. Exons are shown as numbered closed boxes, and loxP sites are shown as closed triangles. (C) Southern blot analysis with probe 2 of BamHI-digested DNA from the tail of mice of the indicated genotypes. The wild-type and floxed alleles give rise to hybridizing fragments of 7.8 and 1.2 kb, respectively. (D) PCR analysis of genomic DNA from the thymus of Fbxw7F/F, Lck-Cre/Fbxw7+/F, and Lck-Cre/Fbxw7F/F mice with the primers Floxed 1 and Floxed 2 (see Materials and methods). The positions of amplified fragments corresponding to wild-type, floxed, and ΔE5 alleles are indicated.