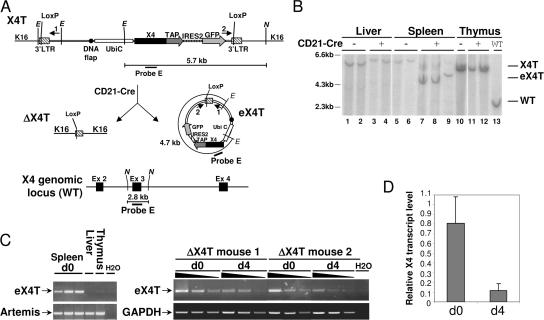
Figure 7.
Production of episomal DNA carrying X4T upon Cre-mediated deletion. (A) Schematic representation of X4T locus before (X4T) and after (ΔX4T) Cre recombination, of episomal form of X4T (eX4T) and of X4 genomic locus (WT). Restriction sites: E, EcoNI; N, NdeI. (B) Southern blot analysis of X4T deletion and eX4T detection, using X4 third exon–specific probe E depicted in A. EcoNI–NdeI–digested DNA from total liver, thymus, or purified splenic mature B cells from X4T X4−/− (CD21-Cre-), ΔX4T X4−/− (CD21-Cre+) and non transgenic (WT) mice, was hybridized with probe E (refer to A for expected sizes of bands). (C) Detection of eX4T by PCR using primers 1 and 2 located in (A), on genomic DNA from the following: (left) purified splenic mature B cells, liver and thymocytes from ΔX4T mice; (right) purified splenic mature B cells from ΔX4T mice before (d0) and after (d4) 4 d of stimulation with LPS and IL-4. eX4T fragment is indicated. Artemis- (left) or GAPDH-specific (right) PCR was used as loading control. (D) Real-time quantitative RT-PCR analysis of X4 transcripts in purified splenic mature B cells from ΔX4T and X4T mice before (d0) and after stimulation with LPS and IL-4 for 4 d (d4). Bars represent the level of X4 transcript in ΔX4T B cells relative to X4T B cells. Results are means of two independent experiments.