Abstract
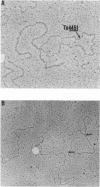
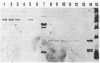
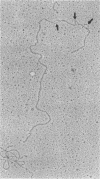
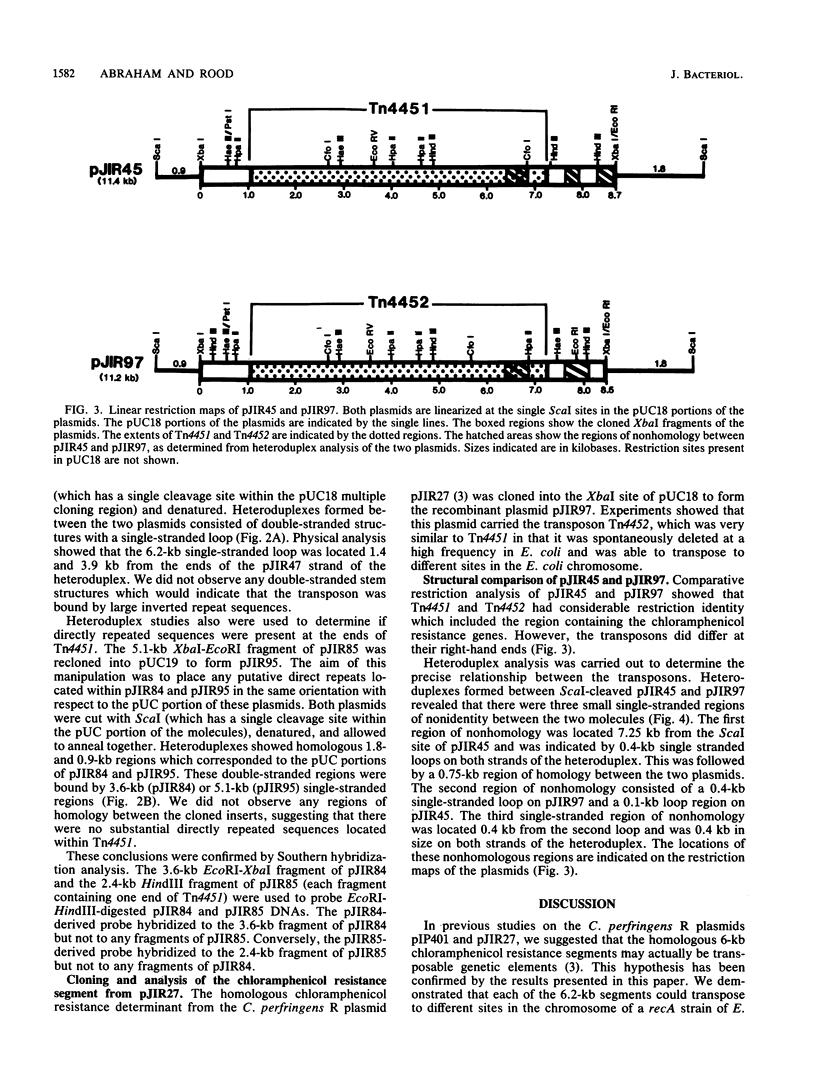
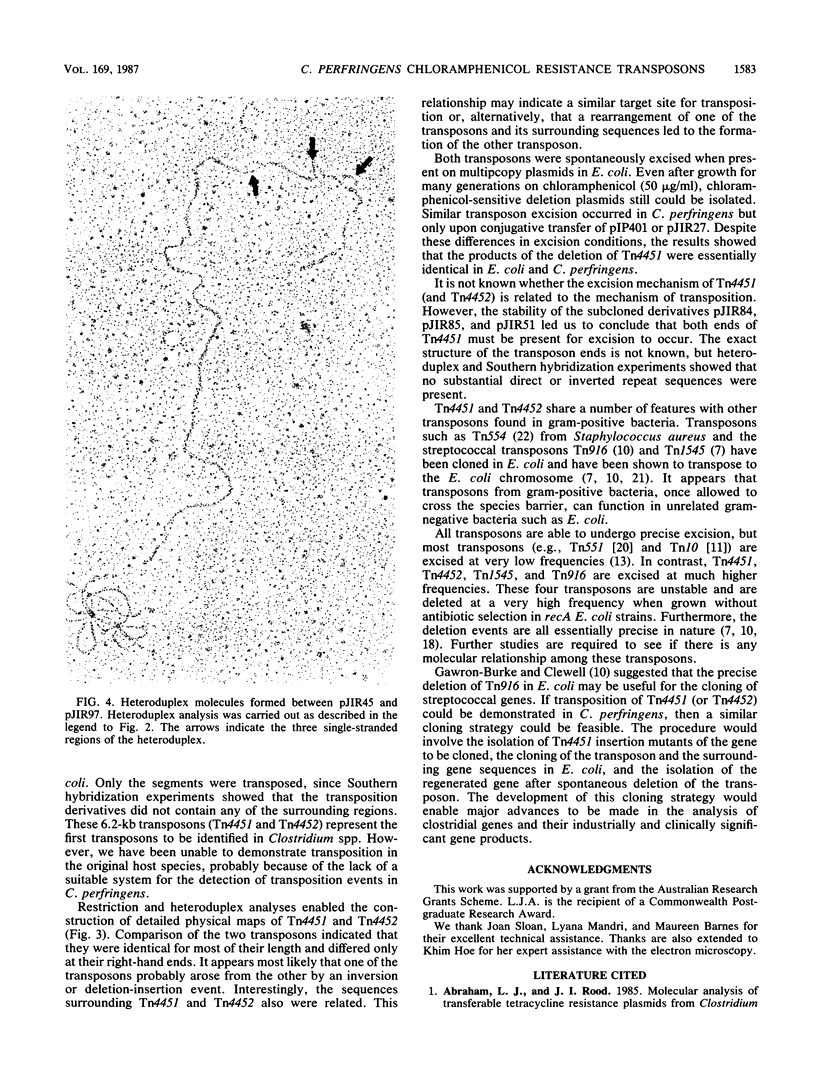
The recombinant plasmids pJIR45 and pJIR97 contain the chloramphenicol resistance determinants derived from the Clostridium perfringens R plasmids pIP401 and pJIR27, respectively. Escherichia coli cultures which harbored these recombinant plasmids rapidly became chloramphenicol sensitive when grown in the absence of chloramphenicol. The loss of resistance was associated with the loss of 6.2-kilobase (kb) segments from both plasmids. Detailed restriction analysis of E. coli- and C. perfringens-derived deletion plasmids indicated that deletion of these segments was essentially precise. Transposition of the 6.2-kb segments was demonstrated by cloning the determinants into a temperature-sensitive plasmid, curing the recombinant plasmids, and selecting chloramphenicol-resistant, plasmid-free clones. Southern hybridization analysis of chromosomal DNA isolated from these recA E. coli clones indicated that the 6.2-kb segments had transposed to different sites on the chromosome. Heteroduplex analysis and restriction mapping indicated that the transposons, Tn4451 (pIP401) and Tn4452 (pJIR27), were closely related and did not contain large inverted or directly repeated sequences. These transposons represent the first transposable elements from the clostridia to be identified and characterized.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham L. J., Rood J. I. Cloning and analysis of the Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid. 1985 May;13(3):155–162. doi: 10.1016/0147-619x(85)90038-1. [DOI] [PubMed] [Google Scholar]
- Abraham L. J., Rood J. I. Molecular analysis of transferable tetracycline resistance plasmids from Clostridium perfringens. J Bacteriol. 1985 Feb;161(2):636–640. doi: 10.1128/jb.161.2.636-640.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham L. J., Wales A. J., Rood J. I. Worldwide distribution of the conjugative Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid. 1985 Jul;14(1):37–46. doi: 10.1016/0147-619x(85)90030-7. [DOI] [PubMed] [Google Scholar]
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brefort G., Magot M., Ionesco H., Sebald M. Characterization and transferability of Clostridium perfringens plasmids. Plasmid. 1977 Nov;1(1):52–66. doi: 10.1016/0147-619x(77)90008-7. [DOI] [PubMed] [Google Scholar]
- Forster A. C., McInnes J. L., Skingle D. C., Symons R. H. Non-radioactive hybridization probes prepared by the chemical labelling of DNA and RNA with a novel reagent, photobiotin. Nucleic Acids Res. 1985 Feb 11;13(3):745–761. doi: 10.1093/nar/13.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984 Jul;159(1):214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I. D., Emmerson P. T. Involvement of recB and recC genes of Escherichia coli in precise transposon excision. J Bacteriol. 1983 Nov;156(2):901–903. doi: 10.1128/jb.156.2.901-903.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kretschmer P. J., Cohen S. N. Selected translocation of plasmid genes: frequency and regional specificity of translocation of the Tn3 element. J Bacteriol. 1977 May;130(2):888–899. doi: 10.1128/jb.130.2.888-899.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge J., Langridge P., Bergquist P. L. Extraction of nucleic acids from agarose gels. Anal Biochem. 1980 Apr;103(2):264–271. doi: 10.1016/0003-2697(80)90266-3. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nida K., Cleary P. P. Insertional inactivation of streptolysin S expression in Streptococcus pyogenes. J Bacteriol. 1983 Sep;155(3):1156–1161. doi: 10.1128/jb.155.3.1156-1161.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Edelman I., Schwesinger M. D., Gruss A. D., Swanson E. C., Pattee P. A. Genetic translocation in Staphylococcus aureus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):400–404. doi: 10.1073/pnas.76.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Murphy E. MLS-resistance determinants in Staphylococcus aureus and their molecular evolution. J Antimicrob Chemother. 1985 Jul;16 (Suppl A):101–110. doi: 10.1093/jac/16.suppl_a.101. [DOI] [PubMed] [Google Scholar]
- Phillips S., Novick R. P. Tn554--a site-specific repressor-controlled transposon in Staphylococcus aureus. Nature. 1979 Mar 29;278(5703):476–478. doi: 10.1038/278476a0. [DOI] [PubMed] [Google Scholar]
- Rood J. I., Maher E. A., Somers E. B., Campos E., Duncan C. L. Isolation and characterization of multiply antibiotic-resistant Clostridum perfringens strains from porcine feces. Antimicrob Agents Chemother. 1978 May;13(5):871–880. doi: 10.1128/aac.13.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebald M., Bréfort M. G. Transfert du plasmide Tétracycline-Chloramphénicol chez Clostridium perfringens. C R Acad Sci Hebd Seances Acad Sci D. 1975 Jul 28;281(4):317–319. [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young I. G., Jaworowski A., Poulis M. I. Amplification of the respiratory NADH dehydrogenase of Escherichia coli by gene cloning. Gene. 1978 Sep;4(1):25–36. doi: 10.1016/0378-1119(78)90012-4. [DOI] [PubMed] [Google Scholar]