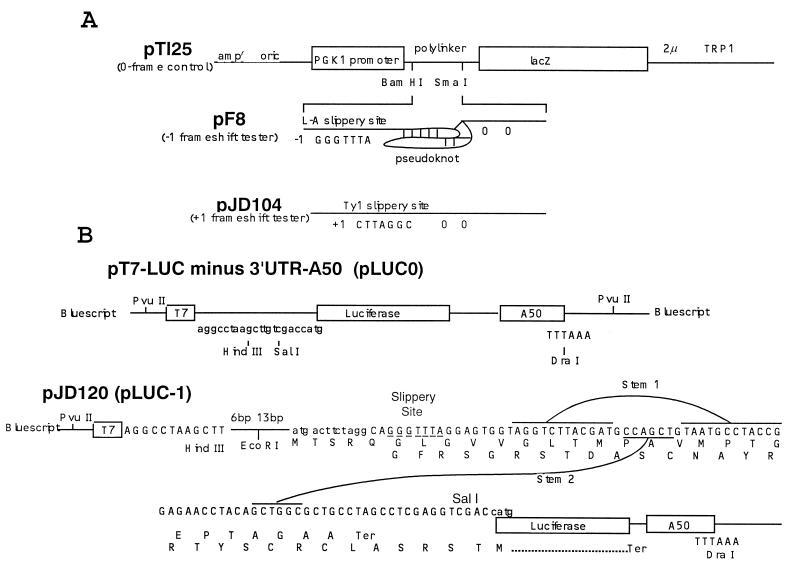
Figure 1.
(A) Vectors used to measure ribosomal frameshift efficiencies in vivo. Plasmid pTI25 and pF8 are as described previously (4), and plasmid pJD104 is as described previously (17). Transcription is driven from the PGK1 promoter and uses the PGK1 translation initiation codon. In pTI25, the bacterial lacZ gene is in the 0-frame with respect to the start site. In plasmid pF8 the lacZ gene is positioned 3′ of the L-A virus frameshift site and in the −1 frame relative to the translation start site. Plasmid pJD104 is derived from pF8 (17) and contains the Ty1 +1 ribosomal frameshift signal 5′ of the lacZ gene. The lacZ gene is inserted in the +1 translation reading frame relative the start of translation. Termination codons in the −1 and +1 frames (pF8 and pJD104, respectively) are located 5′ of the frameshift signals and two 0-frame termination codons are 3′ of the frameshift signal. (B) Vectors to measure ribosomal frameshift efficiencies in vitro. pT7-LUC minus 3′UTR-A50 (LUC0) (18) is the 0-frame reference plasmid. pJD120 (LUC-1) contains the L-A −1 ribosomal frameshift signal cloned into the HindIII/SalI sites of pLUC0. The luciferase coding sequence is in the −1 reading frame with respect to the translational start site such that all luciferase activity must result as a consequence of a −1 ribosomal frameshift event. pLUC0 and pLUC-1 were linearized with DraI and synthetic 7-methyl-Gppp-capped, poly(A)+-tailed transcripts were made using T7 RNA polymerase.