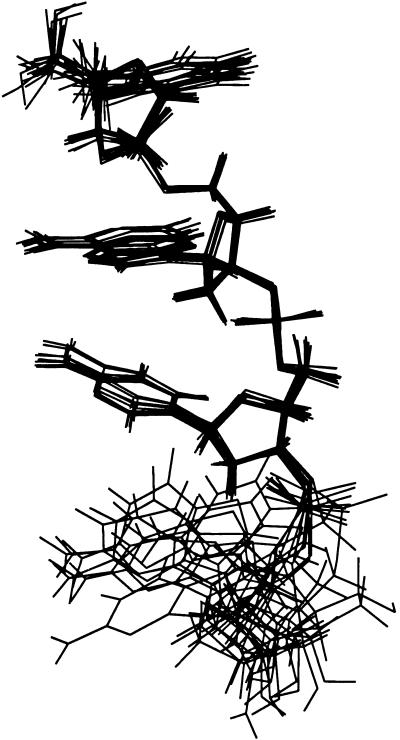
Figure 3.
Superposition of calculated structures of an oligodeoxyribonucleotide, d(TACG). One hundred structures were calculated independently by the use of simulated annealing protocol (x-plor; ref. 26). The 10 lowest energy structures are best fitted at the T-A-C region. Total number of NOE constraints is 59; 39 for intraresidue NOEs and 20 for interresidue NOEs. The root-mean-square deviation of the T-A-C region is 0.30 Å. All residues shows similar deoxyribose-base stacking, whereas the fourth residue (G) is disordered because of few NOE constraints due to signal overlapping. There is no violation to the final structure.