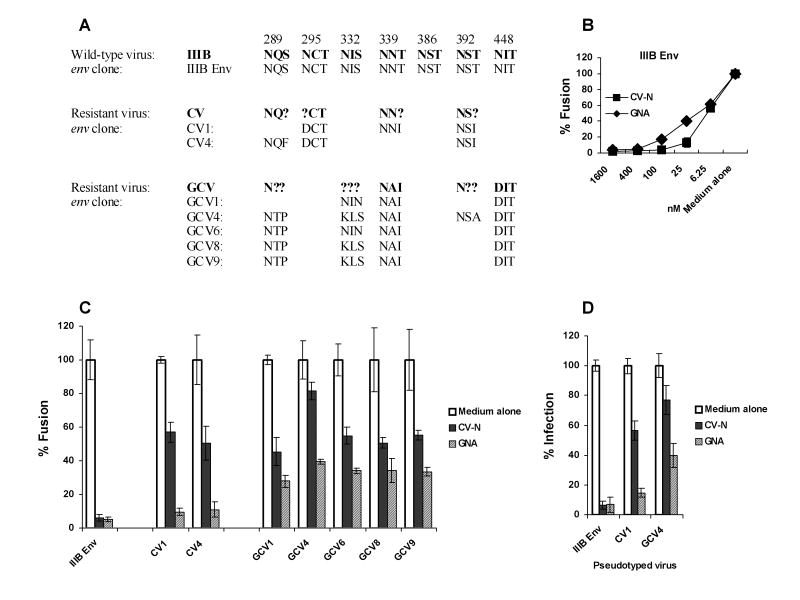
Fig. 2. Genotypic and phenotypic characterization of CV-N resistant env molecular clones.
(A) Alignment of the glycosylation changes in resistant HIV-1 viruses and their env clones. Nonsynonymous sequence polymorphisms in env PCR products, containing both the wild and the mutated amino acids, are indicated by assigning “?” to the position. (B) Fusogenic activity of IIIB Env in the presence of serially diluted CV-N or GNA was determined using the Env mediated cell-cell fusion assay. (C) The fusogenic activity of Env encoded by each env molecular clone in the presence or absence of 100 nM CV-N or 400 nM GNA. The fusogenic activity of each Env in medium alone culture was arbitrarily set to 100%. (D) The infectivity of IIIB Env, CV1 or GCV4 pseudotyped virus in the presence or absence of 100 nM CV-N or 400 nM GNA were assessed in TZM-bl cells. The infectivity of each virus in medium alone culture was arbitrarily set to 100%. Data are representative of 3 independent experiments, with each determination performed in triplicate (mean ± SD).