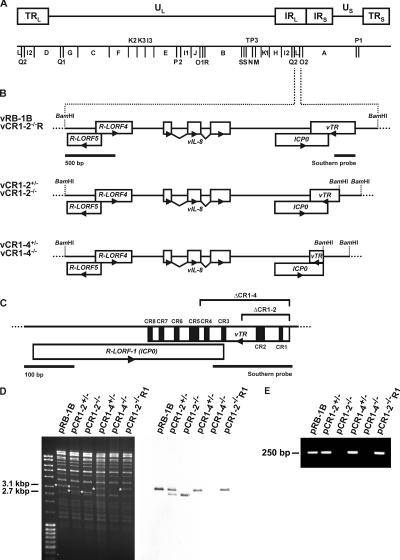
Figure 1.
Genomic structure of vTR deletion and revertant viruses. (A) Schematic presentation of the genomic organization and the BamHI restriction map of MDV. The terminal and internal repeat long regions (TRL, IRL), the unique long region (UL), the internal and terminal repeat short regions (IRS, TRS), and the unique short region (US) are shown. (B) Schematic presentation of the MDV BamHI-L fragment and the genes located therein. The parental (vRB-1B) and mutant viruses lacking either one (vCR1-2+/−, vCR1-4+/−) or both copies of vTR (vCR1-2−/−, vCR1-4−/−) are shown. BamHI restriction sites are given. (C) Detailed schematic presentation of the vTR gene and the putative ICP0 ortholog. The eight CRs of vTR are given in black. Deletion of CRs 1 and 2 (ΔCR1-2) or 1 to 4 (ΔCR1-4) in the genome of vTR mutant viruses is indicated. Also shown is the location of sequences contained in the Southern blot probe. (D) Southern blot analysis of mutant MDV BACs. DNA of pRB-1B and mutant MDV BACs (pCR1-2+/−, pCR1-2−/−, pCR1-4+/−, pCR1-4−/−, and pCR1-2−/−R1) was prepared, digested with BamHI, and separated on a 0.8% agarose gel. Southern blot analysis was performed using a PCR-generated digoxigenin-labeled probe using oligonucleotide primers vTRfw 5′-TGGCGGGTGGAAGGC-3′ and vTRrv 5′-CTGCGGGCGAGGACC-3′. Fragments detected by the vTR probe are indicated by asterisks, and sizes are given. (E) PCR analysis of the mutant BACs. vTR-specific sequences were amplified by using oligonucleotide primers vTRfw and vTRrv, and amplification products were separated on a 1% agarose gel. The size of the specific PCR product is given.