Figure 3.
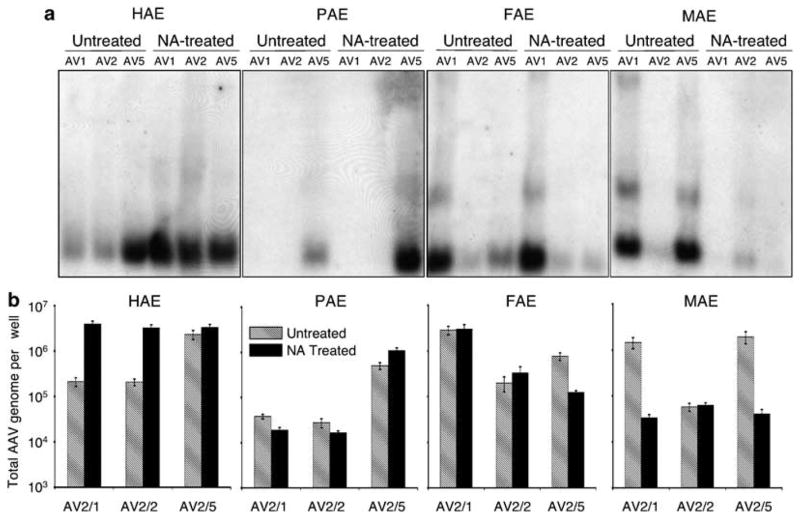
Comparative analysis of N-linked sialic acid-binding requirements for various recombinant adeno-associated virus (rAAV) serotypes. ALI cultures from human (HAE), pig (PAE), ferret (FAE) and mouse (MAE) were treated with neuraminidase (NA-treated) or Ultraser G control medium (untreated) prior to infection. This was followed by application of rAAV luciferase virus to the apical surface at a multiplicity of infection of 2.0 × 103 particles per cell (2 × 109 total particles per well) at 4 °C for 1 h (as described in Figure 2). The infected cultures were washed following virus binding, and cells were harvested for analysis of the viral genome. (a) The extracted viral DNA was detected by Southern blotting using a 32P-labeled luciferase DNA probe as described previously.22,31 (b) Quantitative analyses of rAAV genomes following the above infection protocol used Taqman PCR. Plotted are the means (± s.e.m.) of the numbers of rAAV viral genomes (per well) from three independent experiments (N = 12). The forward primer (5′-TTTTTGAAGCGAAGGTTGTGG-3′), and the reverse primer (5′-CACACACAGTTCGCCTCTTTG-3′) were designed to amplify a 32-bp fragment in the luciferase-coding region of rAAVLuc. The Taqman probe (5′-ATCTGGATACCG GGAAAACGCTGGGCGTTAAT-3′) was tagged with FAM dye (6-carboxyfluorescein) at its 5′-end to serve as the reporter, and with TAMRA dye (6-carboxytetramethylrhodamine) at the 3′-end to serve as the quencher.32 Series dilution of rAAVLuc was used to generate the standard curve, and was only accepted as linear when the slope fell to between −3.74 and −3.32 and the correlation coefficient was above 0.99.32
