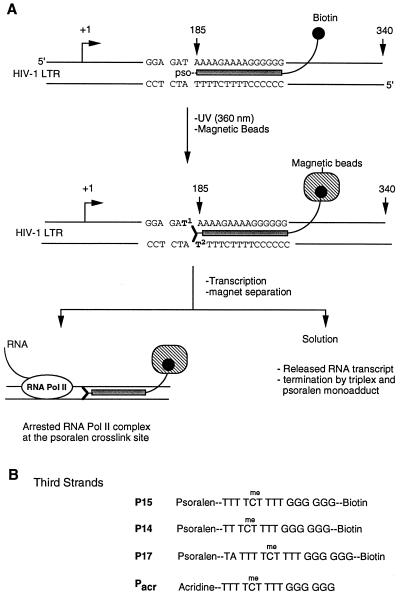
Figure 1.
(A) Experimental design to isolate a homogeneous population of RNA pol II elongation complexes and to study the effects of DNA damage on transcription elongation. A target sequence for triple-helical DNA is inserted into the DNA template. A psoralen and biotin containing oligonucleotide is used to form triplex DNA, and UV irradiation covalently crosslinks psoralen to the template. Psoralen-crosslinked template is immobilized on streptavidin-conjugated magnetic beads, and noncrosslinked DNA is washed away with buffer. Cell-free transcription reactions are performed, and beads are separated from solution by a magnet. The protein composition of the arrested complexes is analyzed by immunoblotting. Different T residues in the target sequence are numbered. Arrows and numbers at +1, 185, and 340 indicate transcriptional start, triplex, and runoff sites, respectively. (B) Structures of third-strand probes used to form triple-helices with the DNA template outlined above. All of these oligonucleotides were modified with psoralen or acridine at their 5′-end. Three probes contained biotin at their 3′-end. C residues in the sequences were substituted with 5-methyl-C to stabilize triplex structures.