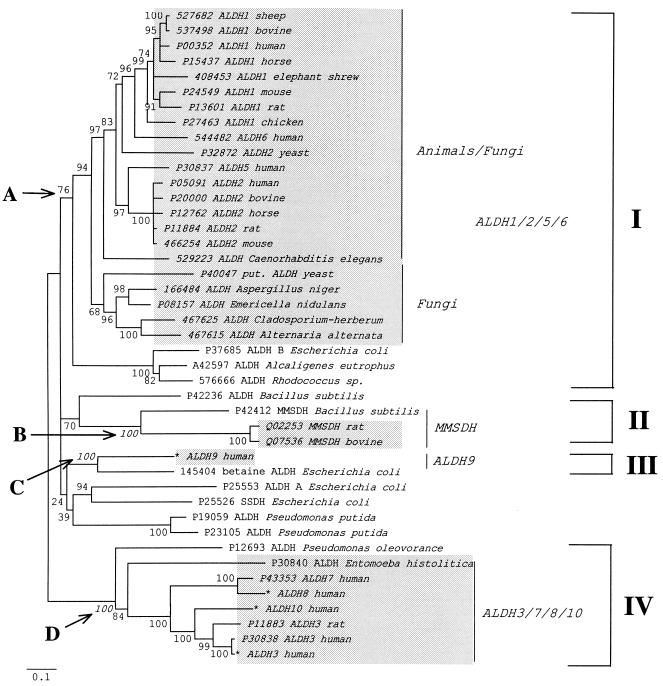
Figure 2.
Neighbor-joining tree (32) generated by mega (33) from 43 ALDH-like protein sequences using the Poisson correction for multiple hits (ref. 34; for ALDH data virtually all currently available corrections for multiple hits give essentially the same tree); the branch lengths are proportional to the number of amino acid substitutions per site. Deletions and insertions were excluded from the analysis. Shaded areas indicate sequences from eukaryotic organisms. We excluded plant ALDHs from the analysis to facilitate interpretation of the resulting phylogeny. Bootstrap P values are shown next to the corresponding interior branches; interior branches that were supported by 20% or less of 500 bootstrap replications (35) were set to zero. The description of each protein sequence includes either the SwissProt or the GenBank accession number (asterisks indicate new sequences) and the protein and species name. A, B, C, and D indicate the interior branches defining four stable clusters of proteins from both eukaryotes and eubacteria. The tree may indicate that at least four ALDH-like genes (ALDH1/2/5/6-like, ALDH3/7/8/10-like, ALDH9-like, and MMSDH-like genes) predated the divergence of eukaryotes and eubacteria. SSDH, succinate-semialdehyde dehydrogenase.