Abstract
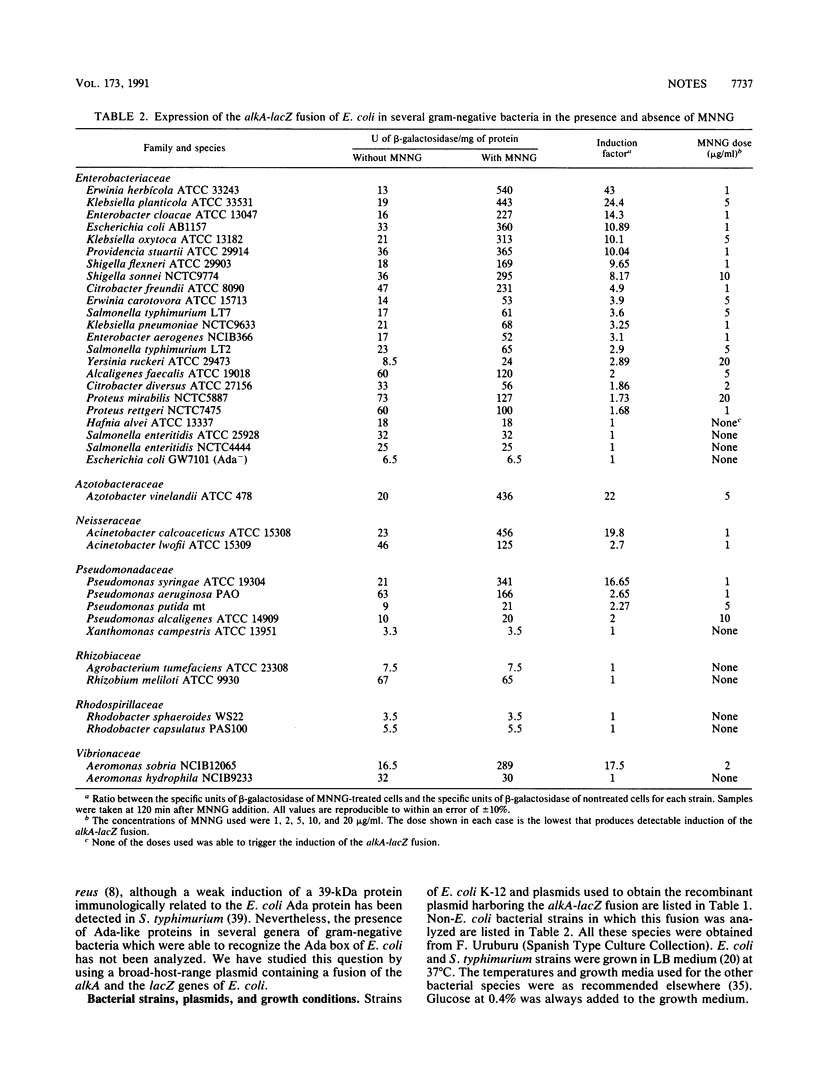
A broad-host-range plasmid containing a fusion of the alkA and lacZ genes of Escherichia coli was introduced into various aerobic and facultative gram-negative bacteria--33 species belonging to 19 genera--to study the induction of expression of the alkA gene by alkylating agents. The bacteria included species of the families Enterobacteriaceae, Pseudomonadaceae, Rhizobiaceae, Vibrionaceae, Neisseriaceae, Rhodospirillaceae, and Azotobacteraceae. Results obtained show that all bacteria tested, except Aeromonas hydrophila, Agrobacterium tumefaciens, Hafnia alvei, Rhizobium meliloti, Salmonella enteritidis, Xanthomonas campestris, and those of the genus Rhodobacter, are able to induce the alkA gene of E. coli in the presence of N-methyl-N'-nitro-N-nitrosoguanidine. All these data indicate that the adaptive response to alkylating agents is present in bacterial species of several families and that the Ada box sequence must be widely conserved.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ather A., Ahmed Z., Riazuddin S. Adaptive response of Micrococcus luteus to alkylating chemicals. Nucleic Acids Res. 1984 Feb 24;12(4):2111–2126. doi: 10.1093/nar/12.4.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R. H., Stonesifer J. Adaptive response and enhancement of N-methyl-N'-nitro-N-nitrosoguanidine mutagenesis by chloramphenicol in Streptomyces fradiae. J Bacteriol. 1985 Nov;164(2):944–946. doi: 10.1128/jb.164.2.944-946.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutibonnes P., Auffray Y. Evidence that N-methyl-N'-nitro-N-nitrosoguanidine induces adaptive response in Bacillus thuringiensis. Mutat Res. 1987 Feb;190(2):83–87. doi: 10.1016/0165-7992(87)90036-4. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Campbell L. A., Yasbin R. E. Mutagenesis of Neisseria gonorrhoeae: absence of error-prone repair. J Bacteriol. 1984 Oct;160(1):288–293. doi: 10.1128/jb.160.1.288-293.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaregola S., D'Ari R., Huisman O. Quantitative evaluation of recA gene expression in Escherichia coli. Mol Gen Genet. 1982;185(3):430–439. doi: 10.1007/BF00334135. [DOI] [PubMed] [Google Scholar]
- Ceccoli J., Rosales N., Goldstein M., Yarosh D. B. Polyclonal antibodies against O6-methylguanine-DNA methyltransferase in adapted bacteria. Mutat Res. 1988 Nov;194(3):219–226. doi: 10.1016/0167-8817(88)90023-5. [DOI] [PubMed] [Google Scholar]
- Demple B., Jacobsson A., Olsson M., Robins P., Lindahl T. Repair of alkylated DNA in Escherichia coli. Physical properties of O6-methylguanine-DNA methyltransferase. J Biol Chem. 1982 Nov 25;257(22):13776–13780. [PubMed] [Google Scholar]
- Demple B., Sedgwick B., Robins P., Totty N., Waterfield M. D., Lindahl T. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(9):2688–2692. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakura A., Morimoto K., Sofuni T., Nohmi T. Cloning and characterization of the Salmonella typhimurium ada gene, which encodes O6-methylguanine-DNA methyltransferase. J Bacteriol. 1991 Jun;173(12):3663–3672. doi: 10.1128/jb.173.12.3663-3672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt P. C., Cooper P. K., Ganesan A. K., Smith C. A. DNA repair in bacteria and mammalian cells. Annu Rev Biochem. 1979;48:783–836. doi: 10.1146/annurev.bi.48.070179.004031. [DOI] [PubMed] [Google Scholar]
- Jeggo P., Defais T. M., Samson L., Schendel P. An adaptive response of E. coli to low levels of alkylating agent: comparison with previously characterised DNA repair pathways. Mol Gen Genet. 1977 Nov 29;157(1):1–9. doi: 10.1007/BF00268680. [DOI] [PubMed] [Google Scholar]
- Kataoka H., Yamamoto Y., Sekiguchi M. A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J Bacteriol. 1983 Mar;153(3):1301–1307. doi: 10.1128/jb.153.3.1301-1307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman A., Walker G. C. A constitutive O6-methylguanine-DNA methyltransferase of Rhizobium meliloti. Mutat Res. 1990 May;235(3):165–169. doi: 10.1016/0921-8777(90)90070-l. [DOI] [PubMed] [Google Scholar]
- Kimball R. F. Further studies on the induction of mutation in Haemophilus influenzae by N-methyl-N'-nitro-N-nitrosoguanidine: lack of an inducible error-free repair system and the effect of exposure medium. Mutat Res. 1980 Aug;72(3):361–372. doi: 10.1016/0027-5107(80)90111-6. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- McCarthy J. G., Edington B. V., Schendel P. F. Inducible repair of phosphotriesters in Escherichia coli. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7380–7384. doi: 10.1073/pnas.80.24.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy T. V., Lindahl T. Methyl phosphotriesters in alkylated DNA are repaired by the Ada regulatory protein of E. coli. Nucleic Acids Res. 1985 Apr 25;13(8):2683–2698. doi: 10.1093/nar/13.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F., Hayashi K., Munakata N. Bacillus subtilis ada operon encodes two DNA alkyltransferases. Nucleic Acids Res. 1990 Sep 25;18(18):5473–5480. doi: 10.1093/nar/18.18.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y., Miyata T., Kondo H., Iwanaga S., Sekiguchi M. Structure and expression of the alkA gene of Escherichia coli involved in adaptive response to alkylating agents. J Biol Chem. 1984 Nov 25;259(22):13730–13736. [PubMed] [Google Scholar]
- Nakabeppu Y., Sekiguchi M. Regulatory mechanisms for induction of synthesis of repair enzymes in response to alkylating agents: ada protein acts as a transcriptional regulator. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6297–6301. doi: 10.1073/pnas.83.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Tokumoto Y., Sakumi K., Koike G., Nakabeppu Y., Sekiguchi M. Expression of the ada gene of Escherichia coli in response to alkylating agents. Identification of transcriptional regulatory elements. J Mol Biol. 1988 Aug 5;202(3):483–494. doi: 10.1016/0022-2836(88)90280-x. [DOI] [PubMed] [Google Scholar]
- Olsson M., Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem. 1980 Nov 25;255(22):10569–10571. [PubMed] [Google Scholar]
- Sakumi K., Sekiguchi M. Regulation of expression of the ada gene controlling the adaptive response. Interactions with the ada promoter of the Ada protein and RNA polymerase. J Mol Biol. 1989 Jan 20;205(2):373–385. doi: 10.1016/0022-2836(89)90348-3. [DOI] [PubMed] [Google Scholar]
- Samson L., Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977 May 19;267(5608):281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- Schneider K., Beck C. F. New expression vectors for identifying and testing signal structures for initiation and termination of transcription. Methods Enzymol. 1987;153:452–461. doi: 10.1016/0076-6879(87)53071-3. [DOI] [PubMed] [Google Scholar]
- Sedgwick B. Genetic mapping of ada and adc mutations affecting the adaptive response of Escherichia coli to alkylating agents. J Bacteriol. 1982 May;150(2):984–988. doi: 10.1128/jb.150.2.984-988.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B. In vitro proteolytic cleavage of the Escherichia coli Ada protein by the ompT gene product. J Bacteriol. 1989 Apr;171(4):2249–2251. doi: 10.1128/jb.171.4.2249-2251.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick S. G., Goodwin P. A. Differences in mutagenic and recombinational DNA repair in enterobacteria. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4172–4176. doi: 10.1073/pnas.82.12.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell D. E., Friedman B. M., Walker G. C. Resistance to alkylation damage in Escherichia coli: role of the Ada protein in induction of the adaptive response. Mutat Res. 1990 Nov-Dec;233(1-2):53–72. doi: 10.1016/0027-5107(90)90151-s. [DOI] [PubMed] [Google Scholar]
- Teo I. A. Proteolytic processing of the Ada protein that repairs DNA O6-methylguanine residues in E. coli. Mutat Res. 1987 Mar;183(2):123–127. doi: 10.1016/0167-8817(87)90054-x. [DOI] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Demple B., Li B., Lindahl T. Induction of resistance to alkylating agents in E. coli: the ada+ gene product serves both as a regulatory protein and as an enzyme for repair of mutagenic damage. EMBO J. 1984 Sep;3(9):2151–2157. doi: 10.1002/j.1460-2075.1984.tb02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Kilpatrick M. W., McCarthy T. V., Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986 Apr 25;45(2):315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- Vaughan P., Sedgwick B. A weak adaptive response to alkylation damage in Salmonella typhimurium. J Bacteriol. 1991 Jun;173(12):3656–3662. doi: 10.1128/jb.173.12.3656-3662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vericat J. A., Barbé J. Adaptive response to simple alkylating agents in the phototrophic bacteria Rhodobacter capsulatus and R.sphaeroides. Mutagenesis. 1988 Mar;3(2):165–168. doi: 10.1093/mutage/3.2.165. [DOI] [PubMed] [Google Scholar]
- Volkert M. R. Adaptive response of Escherichia coli to alkylation damage. Environ Mol Mutagen. 1988;11(2):241–255. doi: 10.1002/em.2850110210. [DOI] [PubMed] [Google Scholar]
- Volkert M. R., Nguyen D. C., Beard K. C. Escherichia coli gene induction by alkylation treatment. Genetics. 1986 Jan;112(1):11–26. doi: 10.1093/genetics/112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinfeld M., Drake A. F., Saunders J. K., Paterson M. C. Stereospecific removal of methyl phosphotriesters from DNA by an Escherichia coli ada+ extract. Nucleic Acids Res. 1985 Oct 11;13(19):7067–7077. doi: 10.1093/nar/13.19.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Katsuki M., Sekiguchi M., Otsuji N. Escherichia coli gene that controls sensitivity to alkylating agents. J Bacteriol. 1978 Jul;135(1):144–152. doi: 10.1128/jb.135.1.144-152.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]


