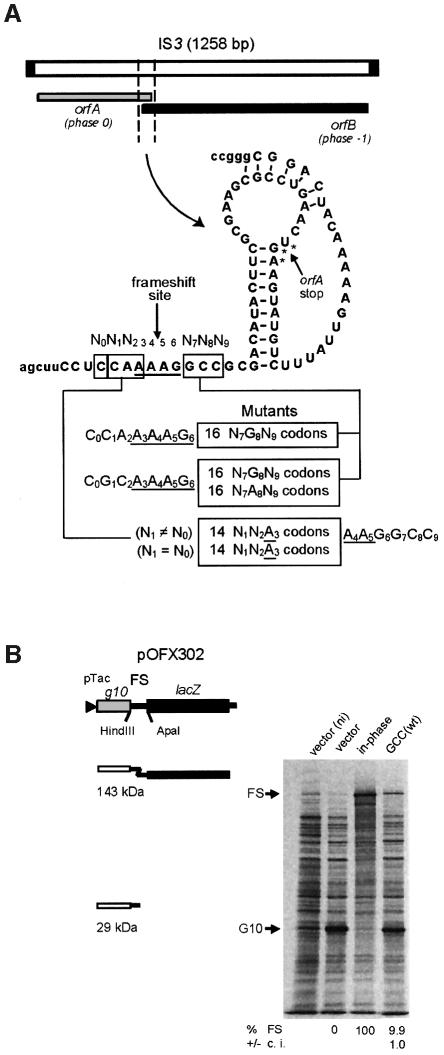
Fig. 1. The IS3 frameshift region, its various derivatives and the plasmid reporter system. (A) The segment of IS3 shown and the derived mutants were cloned in the pOFX302 plasmid. (B) Frameshifting efficiency was determined by protein labeling with [35S]methionine. The results obtained with two control strains (one with an in-phase construct, giving the theoretical 100% frameshifting value, and the other containing the vector plasmid, 0% frameshifting value), and with the IS3 ‘wild type’ region (wt) are shown. One culture of the vector- containing strain was labeled in the absence of IPTG (ni). The position of the product from normal translation (G10) or frameshifting (FS) is indicated. The calculated level of frameshifting and the 95% confidence interval (c.i.) are indicated below the relevant lanes. In the natural IS3 frameshift region an AUG codon (frame –1) overlapping the UGA stop codon of orfA (frame 0) is used to initiate synthesis of the OrfB protein. This AUG codon was changed to CUG in order to prevent initiation without interfering with frameshifting (Sekine et al., 1994).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.