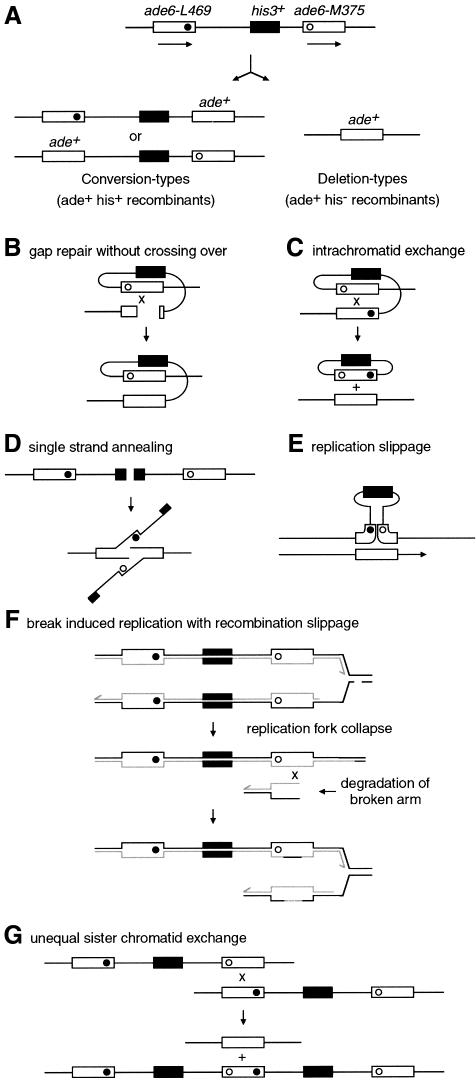
Fig. 5. A non-tandem direct repeat of heteroalleles for measuring recombination. (A) Schematic of intrachromosomal recombination substrate and recombinant products. Solid and open circles represent the ade6-L469 and ade6-M375 mutations, respectively. (B–G) Some of the possible mechanisms of deletion- and conversion-type recombinant formation. (B) DSB/gap repair involving intrachromatid recombination. If the recombination intermediate is resolved to give a non-crossover product, then this can generate an ade+ his+ conversion type. (C) Intrachromatid exchange between the direct repeats generates a single copy of the gene in the chromosome and an excised circle bearing the second copy together with the his+ gene. (D) Single strand annealing (SSA) can repair a DSB between two direct repeats by resection of the broken ends to expose two complementary single strands that anneal. Repair involves removal of the non-homologous 3′ ends. (E) Replication slippage involving detachment of a nascent strand and mispairing within repeats upon reattachment. (F) Break-induced replication (BIR). This may occur when a replication fork collapses after encountering a single strand break in the template DNA. Recombination can restore the fork by reattaching the broken end. If the broken arm of the fork is degraded, then recombination may occur between non-equivalent repeats, resulting in the formation of deletion-type recombinants. In this version of the BIR model, the reformation of the replication fork is accompanied by the formation and resolution of an HJ (not shown). In other versions, replication is envisaged to proceed via a migrating bubble or D-loop (Paques and Haber, 1999). (G) Unequal sister chromatid exchange can result in a deletion on one chromatid and a triplication on the other chromatid. Such unequal or slipped recombination can generate conversion-type recombinants if recombination intermediates are processed without crossing over.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.