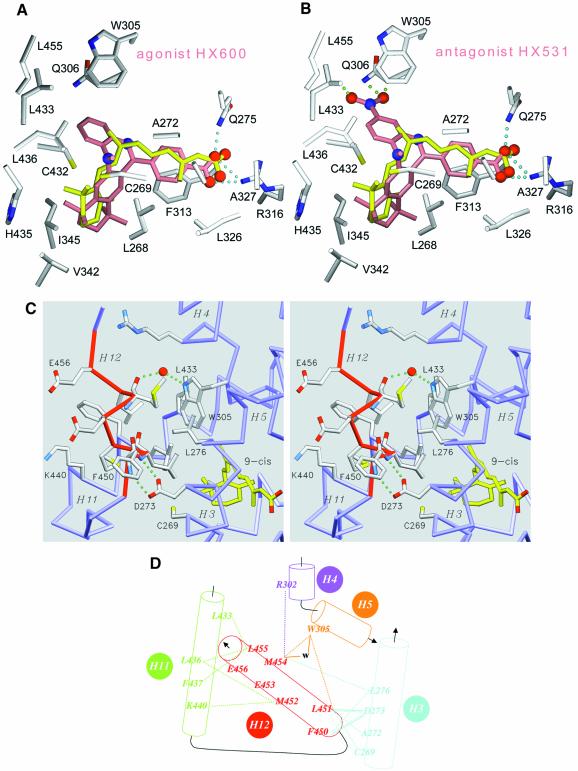
Fig. 3. The agonist conformation of transactivation helix H12 in the holo form. Docking of retinoid agonist (HX630) and antagonist (HX531) in hRXRα LBP. (A and B) Agonist (HX600) and antagonist (HX531) compounds docked in the LBP of hRXRα. Protein atoms are coloured in grey for carbon, blue for nitrogen, red for oxygen and yellow for sulfur. The oxygen and nitrogen atoms of docked compounds are depicted as red and blue spheres, respectively. 9-cRA is coloured in yellow, whereas docked ligands are colored in salmon. Cyan dotted lines represent the structurally conserved hydrogen interaction between the carboxylic moiety of ligands and residues Arg316 and Gln275 of the protein. Green dotted lines underline steric clashes through close interatomic contacts between ligand and protein atoms (the distance between consecutive dots is 0.5 Å). (C) Detailed stereoview of helix H12 contacts showing the exposed glutamic residues Glu453 and Glu456 involved in transactivation and the interactions stabilizing helix H12 in its agonist position. Helix H12 is depicted in red. 9-cRA ligand atoms are coloured in yellow for carbon and red for oxygen, respectively. Protein atoms are coloured in grey for carbon, blue for nitrogen, red for oxygen and yellow for sulfur. The protein backbone is coloured in blue. A water molecule is drawn as a red sphere and hydrogen bonds are depicted as green dotted lines. For the sake of clarity only a few side chains are labelled. (D) Schematic drawing of interactions stabilizing H12 in its agonist conformation. van der Waals interactions and hydrogen bonds are represented as dotted and continuous lines, respectively. A water molecule is referred to as w.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.