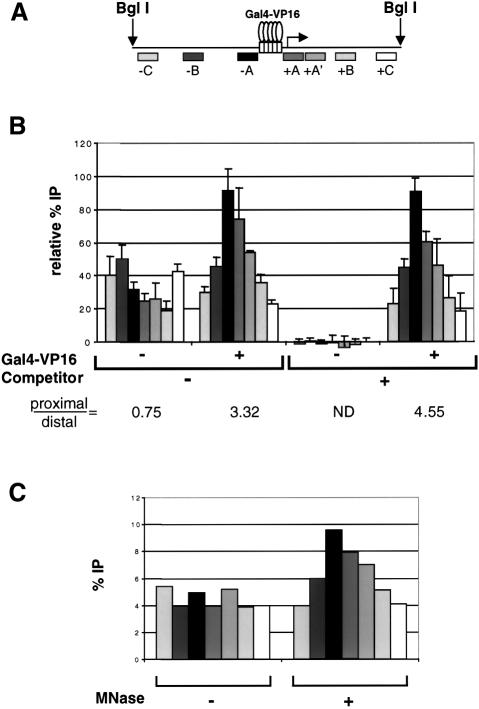
Fig. 5. Gal4-VP16 directs the HAT activity of the SAGA complex to promoter-proximal nucleosomes. The experimental set-up for these ‘scanning ChIPS’ is similar to that used in Figure 4: the template was incubated in the absence (–) or presence (+) of Gal4-VP16, and competitor chromatin was added where indicated. Next, the reactions were incubated with SAGA and acetyl-CoA (or mock acetylated in the absence of HAT complex), MNase digested and immunoprecipitated with anti-acetylated H3 antibody. DNA was extracted from the bound and unbound fractions and slot-blotted. The membranes were hybridized successively with a series of short probes (between 250 and 300 bp) that scan the length of the template, generated by PCR and labeled by primer extension from random hexanucleotides (Boehringer Mannheim). (A) Diagram showing the localization of the different probes when the plasmid is digested with BglI. (B) Average values and standard deviation of normalized data from three repeats of the experiment. The background signal (–HAT) was subtracted from the values obtained in the presence of SAGA for each condition. The numbers under the graph show the ratio of proximal (average of +A and –A) versus distal (average of +C and –C) signal for each condition tested. ND, not determined. (C) The reconstituted array was pre-incubated with Gal4-VP16 and competitor chromatin was added. Then, the template was acetylated by SAGA in the presence of acetyl-CoA, reactions were digested with MNase (+) or mock-digested (–), and the immunoprecipitation and slot blot were carried out as described above.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.