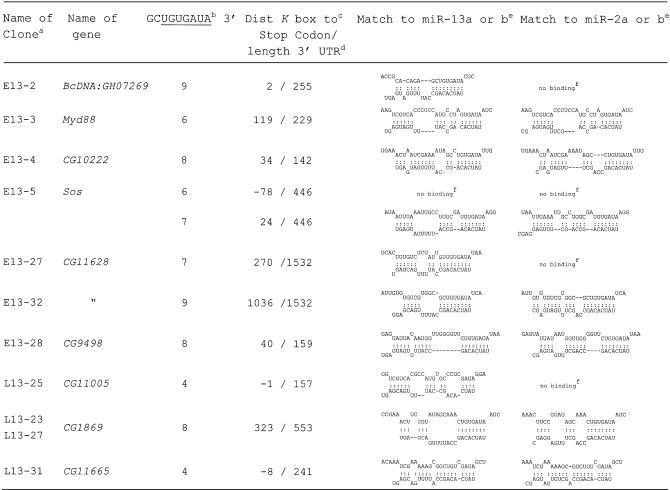
Table 1. Identified Drosophila genes, their 3′ UTR motifs and potential interaction with miR-13 and miR-2.
aClones originating from an early and late cDNA library are denoted with E13 and L13, respectively.
bNumber of nucleotides matching to the 3′ end of the outlined 9 nt sequence element, wherein the K box is underlined.
cNumber of nucleotides between the last nucleotide of the stop codon and the 3′ terminal A of the K box and matching to the 5′ U of miR-13. Negative numbers indicate that the miR-13 target site is located upstream of the stop codon or overlapping with it (CG1105).
dThe given length of the 3′ UTR is based on sequence data from the cDNA clones; it indicates the number of nucleotides between the stop codon and poly(A).
eThe structures have been calculated with the Mfold program (38) by artificially joining the two RNAs via a oligo A or C; the sequences of miR-13a and miR-13b refer to table 1 of Lagos-Quintana et al. (1).
fNo binding, only interaction of the two RNAs via the K box.