Figure 1.
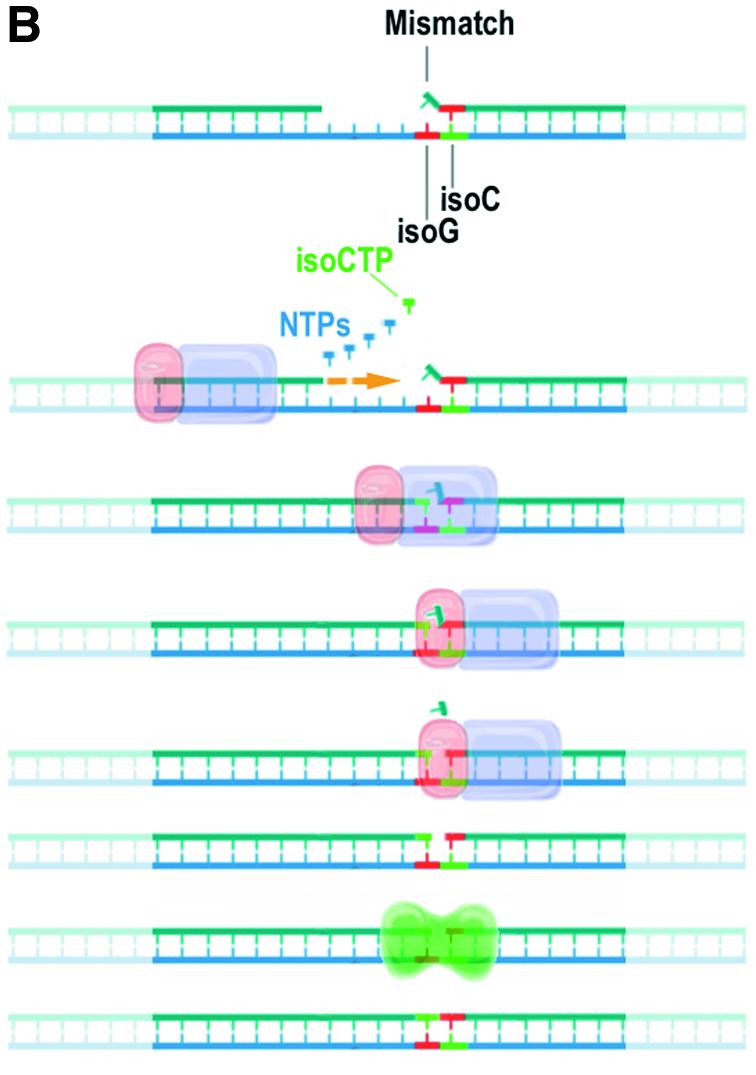
(A) Alternative bases that are structurally similar to standard nucleobases but have altered base-pairing specificity. The pattern of hydrogen bond acceptors and donors (indicated by arrows) is reversed for the non-standard base pair between 5-methyl isocytosine (isoC) and isoguanosine (isoG) (right) compared with a standard cytosine (C) guanosine (G) base pair (left). (B) Mismatch repair model of a template containing consecutive non-standard base pairs. The polymerase domain (purple) of a bacterial DNA polymerase catalyzes repair DNA synthesis by incorporation of nucleoside triphosphates. The 5′ nuclease domain (pink) of the polymerase excises a standard base (blue) mismatched with a non-natural base (isoG, red) in the template. DNA ligase (green) completes repair by formation of a covalent bond between isoC (green) and isoG (red).

