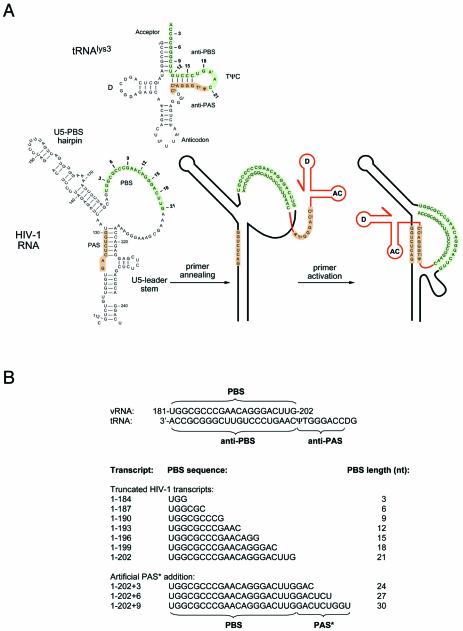
Figure 1.
(A) Scheme of the tRNAlys3 primer and part of the HIV-1 RNA genome. Marked are the PBS and PAS sequences in the HIV-1 untranslated leader RNA and the antiPBS and antiPAS sequences in the tRNAlys3 primer. In the secondary structure model of the HIV-1 RNA we marked the PAS element, U5-top hairpin, PBS and the U5-leader stem. Within the PBS, the positions corresponding to the 3′ terminus of truncated transcripts used in this study are indicated in bold. Numbers indicate the length of the PBS for the corresponding transcript. (B) PBS sequence for the truncated and extended HIV-1 transcripts used in this study. The PBS length in this set of transcripts ranges from 3 to 30 nt. The set includes three transcripts that progressively have an extension of the PBS/tRNA complementarity, such that an artificial PAS* sequence is fused directly downstream of the PBS.