Figure 1.
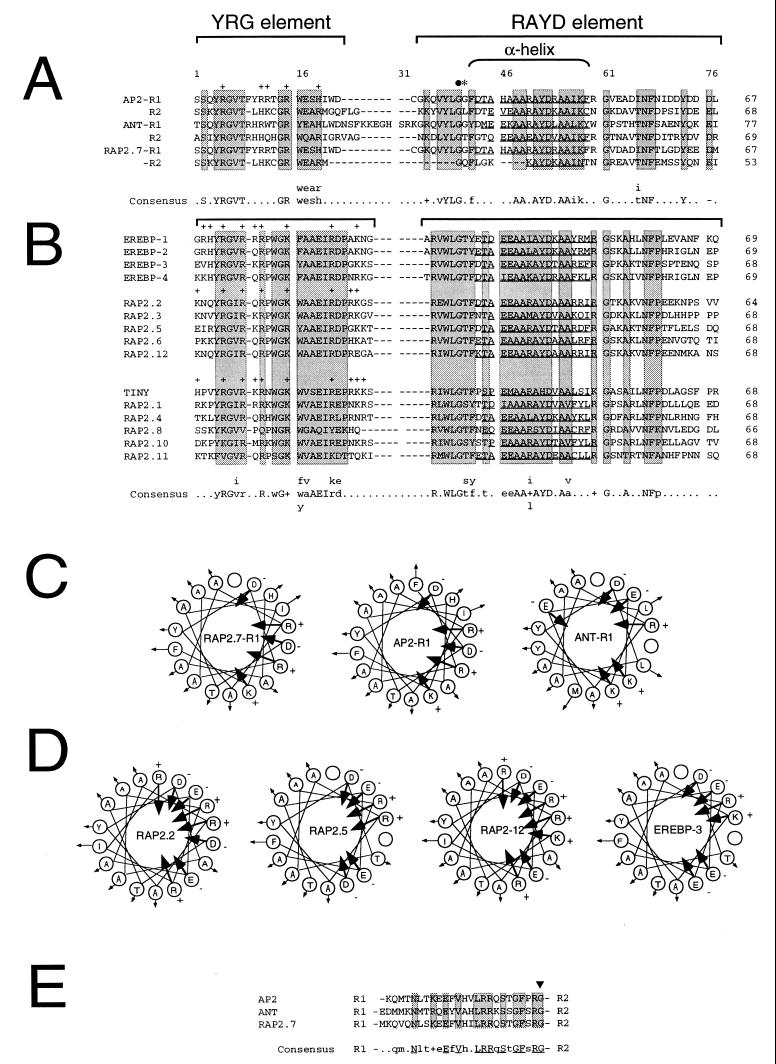
AP2 domain sequence and structure. Arabidopsis RAP2 amino acid sequences were determined and compared as described. Gene names are shown to the left of each sequence. The number of amino acid residues within each AP2 domain is shown to the right. Sequence gaps were introduced to maximize sequence alignments. The position of amino acid residues and sequence gaps within the AP2 domain alignments are numbered 1–77 for reference. The location of the conserved YRG and RAYD elements are indicated by brackets. Shaded boxes highlight regions of sequence similarity. Positively charged amino acids within the YRG element are indicated by + signs above the residues. The location of the 18-amino acid core region that is predicted to form an amphipathic α-helix in AP2 (9) is indicated by a bracket. Residues within the RAYD element of each AP2 domain that are predicted to form an amphipathic α-helix are underlined. (A) AP2-like AP2 domains. Amino acid sequence alignment between the AP2 domain repeats R1 and R2 contained within AP2 (9), ANT (14, 15), and RAP2.7 is shown. Brackets above the sequences designate the conserved YRG and RAYD blocks described above. The filled circle and asterisk indicate the positions of the ap2-1, and ap2-5 mutations, respectively (9). Amino acid residues that constitute a consensus AP2 domain motif for AP2, ANT, and RAP2.7 is shown below the alignment with invariant residues shown capitalized. The GenBank accession number for AP2 is U12546. (B) EREBP-like AP2 domains. Amino acid sequence alignment between the AP2 domains contained within the tobacco EREBPs and the Arabidopsis EREBP-like RAP2 proteins is shown. GenBank accession numbers for EREBP-1, EREBP-2, EREBP-3, and EREBP-4 are D38123, D38126, D38124, and D38125, respectively. (C) Schematic diagrams of the putative RAP2.7-R1, AP2-R1, and ANT-R1 amphipathic α-helices. Amino acid residues within the RAP2.7-R1, AP2-R1, and ANT-R1 motifs shown underlined in A that are predicted to form amphipathic α-helices are schematically displayed with residues rotating clockwise by 100° per residue to form helical structures. Arrows directed toward or away from the center of the helical wheel diagrams indicate the negative or positive degree of hydrophobicity as defined by Jones et al. (33). Positively and negatively charged amino acid residues are designated by + and − signs, respectively. (D) Schematic diagrams of the putative RAP2.2, RAP2.5, RAP2-12, and EREBP-3 amphipathic α-helices. Amino acid residues within the RAP2.2, RAP2.5, RAP2-12, and EREBP-3 motifs shown underlined in B that are predicted to form amphipathic α-helices are schematically displayed as described in C. (E) Sequence alignment between the 25–26 amino acid linker regions in AP2, ANT, and RAP2.7. R1 and R2 designate the positions of the R1 and R2 repeats within AP2, ANT, and RAP2.7 relative to the linker region sequences. Boxes designate invariant residues within the conserved linker regions. Amino acid residues that constitute a consensus linker region motif for AP2, ANT, and RAP2.7 are shown below the alignment with invariant residues shown capitalized. The arrowhead indicates the position of the ant-3 mutation described by Klucher et al. (15).