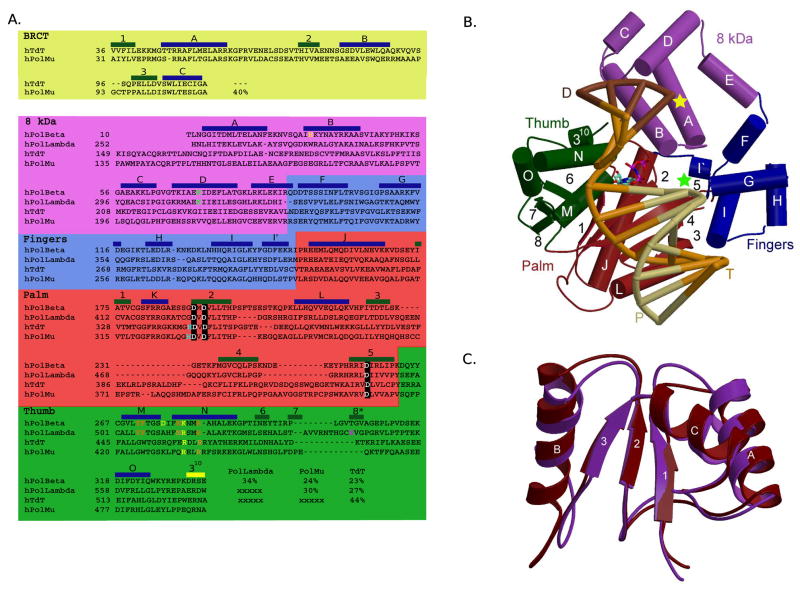
Figure 1.
Mammalian Family X polymerases. (A) Structure-based primary sequence alignment. Catalytic aspartates, boxed in black; residues involved in minor groove interactions, orange; residues involved in stacking interactions with nucleotides, yellow; dRP lyase nucleophile, green; active site histidines (Pol μ H329, TdT H342), cyan; residue putatively stabilizing frameshift intermediate, magenta. Secondary structural elements are blue (α-helices), green (β-strands), and yellow (310 helix) boxes. (B) Domain organization. α-helices are depicted as cylinders, and labeled alphabetically. β-strands are shown as directional arrows, labeled numerically. The bound DNA duplex is shown in the binding cleft: template (T), orange; upstream primer (P), khaki; and downstream primer (D), brown. The position of the 3′-OH and downstream 5′-phosphate are marked with green and yellow stars, respectively. The incoming nucleotide is cyan. The ternary complex of Pol β (PDB code 2FMS) was used for this illustration. (C) NMR solution structure of the Pol μ (maroon) and TdT (purple) BRCT domains. α-helices are labeled alphabetically and β-strands numerically. Numbering only applies within the BRCT domains.