Figure 4.
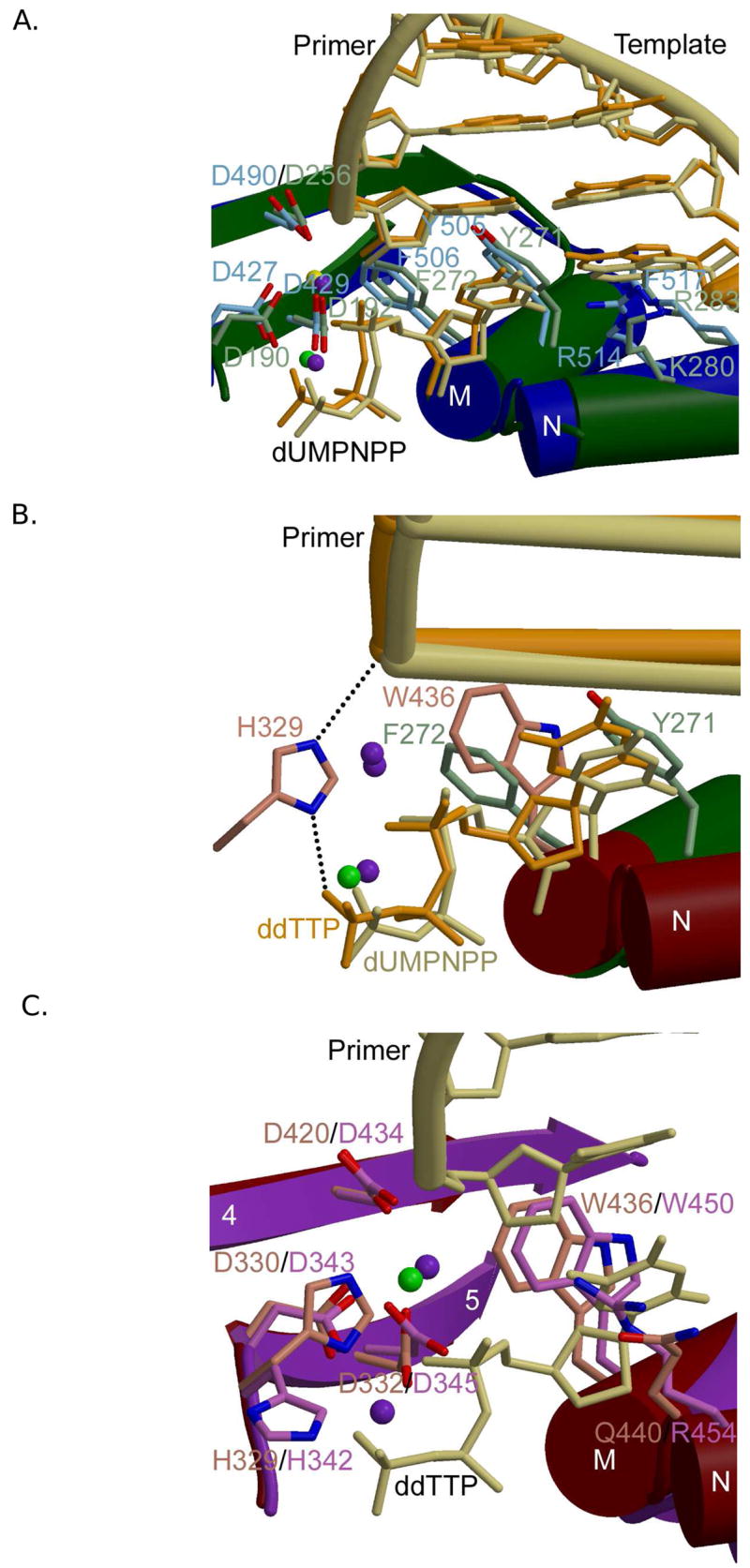
Family X polymerase active site organization. (A) Superposition of ternary complexes of Pol β (PDB code 2FMS; protein, green; DNA, khaki) and Pol λ (PDB code 2PFO; protein, blue; DNA, orange). Protein side chains from Pol β are drawn in light green, while those for Pol λ are light blue. (B) Superposition of the ternary complexes of Pol β (PDB code 2FMS; protein, green; DNA, khaki) and Pol μ (PDB code 2IHM; protein, maroon; DNA, orange). Protein side chains from Pol β are drawn in light green, while those for Pol μ are pink. Putative hydrogen bonding interactions between Pol μ H329 and the primer terminal phosphate or the γ-phosphate of the incoming nucleotide are shown as black dashed lines. Magnesium and sodium ions are shown as purple and green spheres, respectively. (C) Superposition of ternary complexes of Pol μ (PDB code 2IHM; protein, maroon; DNA, khaki) and TdT apoprotein (PDB code 1JMS; protein, purple). Protein side chains from Pol μ are drawn in pink, while those for TdT are light purple. The incoming nucleotide from the structure of Pol μ is drawn in khaki. Magnesium and sodium ions are shown as purple and green spheres, respectively.
