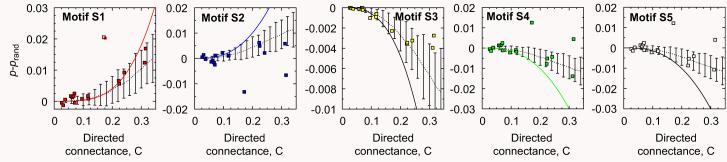
Fig. 8.
Differences between actual appearances of motifs and the corresponding randomized food webs for 17 empirical food webs (symbols) as compared to the analytical predictions for the generalized cascade model (solid line) and the random model (dashed lines). Numerical simulations for the generalized cascade model with S = 50 are shown by the dotted line, where the error bars are two standard deviations. Motifs S1 and S2 are typically over-represented and motifs S3–S5 are under-represented, in agreement with the qualitative predictions of the model. The two noticeable deviations correspond to Bridge Brook (C = 0:17) and Skipwith Pond (C = 0:32). Quantitatively, the analytical curves generally overestimate the differences at larger values of C for both the empirical values and the numerical simulations of the generalized cascade model.