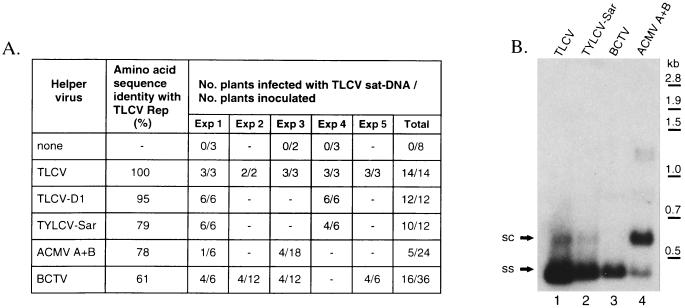
Figure 8.
Replication of TLCV sat-DNA is supported by different geminiviruses encoding heterologous replication-associated proteins. Datura plants were agroinoculated with a dimeric clone of TLCV sat-DNA alone or together with infectious constructs of different helper geminiviruses: TLCV, TLCV-D1 strain, TYLCV-Sardinian strain, ACMV A + B or BCTV. All plants inoculated with these viruses developed disease symptoms. A, Summary of satellite infectivity experiments as determined by dot–blot analysis using a TLCV sat-DNA probe. Amino acid sequence identity between the TLCV Rep and the protein homologue of the other geminiviruses was calculated using GAP. B, Southern blot of total nucleic acid extracts of plants identified as positive for satellite DNA. Replicative forms of satellite DNA were detected with a TLCV sat-DNA probe. Due to the large variation in satellite DNA levels obtained with the different helper viruses, the relative loadings of total nucleic acid per lane were adjusted as follows: lanes 1 and 2 (0.75 μg); lane 3 (0.31 μg); lane 4 (5 μg) representing relative loadings of 1:1:16:66 for lanes 1–4, respectively. TLCV sat-DNA forms are labeled as in Fig. 2.