Fig. 1.
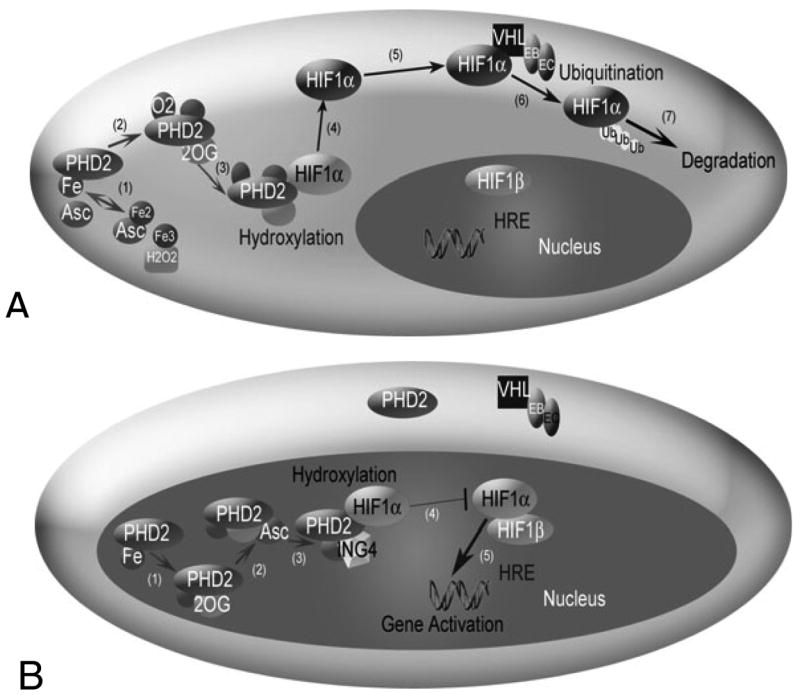
The HIF1 pathway in normoxia (A) and hypoxia (B). (A) HIF1α hydroxylation and degradation in the presence of oxygen involves: (1) the independent oxidation-reduction reactions of ascorbate (Asc) and iron (Fe); (2) and (3) prolyl hydroxlyase 2 (PHD2) binding to Fe, 2-oxoglutarate (2OG), and O2; (4) PHD2 hydroxylation of HIF1α; (5) unbound hydroxylated HIF1α moving in the cell cytoplasm; (6) the von Hippel Lindau (VHL)-Elongin B (EB)-Elongin C (EC) complex ubiquitylating HIF1α; and (7) HIF1α degradation. A change in shading of HIF1α indicates addition of a hydroxyl group. (B) In hypoxia, HIF1α enters the nucleus, where hydroxylation, but no degradation occurs. (1) and (2) PHD2 binding to Fe, 2OG and Asc, but not O2. (3) The protein inhibitor of growth 4 (ING4) binding to PHD2 may regulate HIF1α transcriptional activity and (4) block HIF1α-HIF1β binding. When HIF1α-HIF1β binding occurs, the HIF1 dimer can transcriptionally activate genes at the hypoxia response element (HRE) site.
