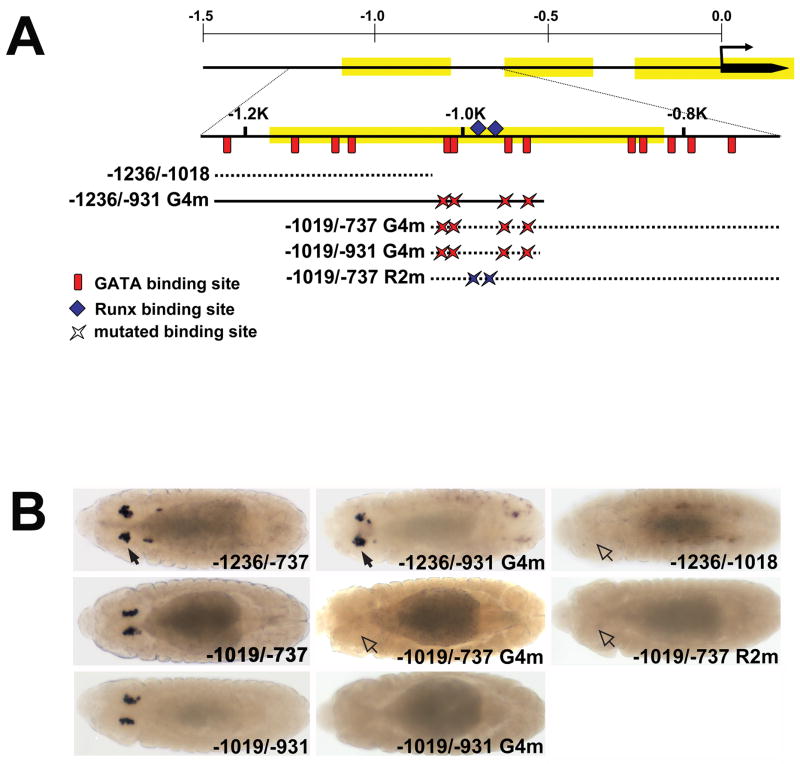
Fig. 2. Clustered GATA and RUNX motifs are required for lz crystal cell CRM activity.
(A) Schematic showing fragments with mutated GATA and RUNX motifs used to evaluate lz-lacZ CRM activity. The 1.5 kb region upstream of the lz transcription start site is depicted with the evolutionarily conserved regions highlighted in yellow. Beneath the map of the 1.5 kb region is an expanded map of the region from −1250 to −727. Red vertical lines show the relative positions of the 13 GATA motifs between positions −1207 and −768. The blue diamonds mark the positions of the two RUNX motifs located at −995 and −977. Mutations in the four GATA motifs, between positions −1010 and −947, are designated G4m, whereas the mutations in the two RUNX motifs are designated R2m. The mutated fragments are numbered and positioned relative to the transcription start site, with the suffix G4m or R2m to indicate disruption of the GATA or RUNX motifs, respectively. Solid line represents a fragment with crystal cell activity, whereas dashed lines represent fragments lacking crystal cell activity. (B) Comparison of wild-type and GATA or RUNX mutant CRMs. Dorsal views of stage 13 embryos. lz-lacZ strains are indicated in the lower right hand corner of each panel. Closed arrows mark activity in crystal cells; open arrows mark lack of activity. Abbreviations: G4m, GATA Core mutants; R2m, RUNX mutants.