Abstract
Promoters control the expression of genes in response to one or more transcription factors (TFs). The architecture of a promoter is the arrangement and type of binding sites within it. To understand natural genetic circuits and to design promoters for synthetic biology, it is essential to understand the relationship between promoter function and architecture. We constructed a combinatorial library of random promoter architectures. We characterized 288 promoters in Escherichia coli, each containing up to three inputs from four different TFs. The library design allowed for multiple −10 and −35 boxes, and we observed varied promoter strength over five decades. To further analyze the functional repertoire, we defined a representation of promoter function in terms of regulatory range, logic type, and symmetry. Using these results, we identified heuristic rules for programming gene expression with combinatorial promoters.
Keywords: combinatorial promoter, logic gate, signal integration, synthetic biology, transcription regulation
Introduction
In many promoters gene expression is regulated in response to two or more transcription factors (TFs). A classic example is the lac operon, where promoter activity depends on both the repressor LacI (Jacob and Monod, 1961) and the activator CRP (Zubay et al, 1970). Such combinatorial regulation of gene expression underlies diverse cellular programs (Ptashne, 2005), including responses to environmental conditions (Ligr et al, 2006) and multicellular development. Combinatorial promoters with multiple TF binding sites, or operators, can facilitate the integration of multiple signals. For example, a synthetic combinatorial promoter responding to LuxR and λ cI was recently used to construct a genetic pulse generator (Basu et al, 2004), a band-pass filter, and a bulls-eye pattern formation system (Basu et al, 2005). Furthermore, circuits containing combinatorial promoters are predicted to generate robust oscillations (Hasty et al, 2002; Atkinson et al, 2003) or to create sign-sensitive filters, signal averaging, and response acceleration or delay (Mangan and Alon, 2003).
Bacterial promoters typically occupy a region of 100 bp or less, surrounding the start site (+1) of transcription, approximately from positions −75 to +25. This sequence includes the primary binding sites for the polymerase, the −10 and −35 boxes (Hawley and McClure, 1983), additional upstream (Chan and Busby, 1989; Ross et al, 1993) and downstream (Kammerer et al, 1986; Haugen et al, 2006) regulatory sequences, along with operators for activator and/or repressor TFs (Busby and Ebright, 1994; Browning and Busby, 2004). Operators within this region enable bound TFs to directly contact and recruit the polymerase (activation) or to sterically block polymerase contact with the −10 and −35 boxes (repression). The type and arrangement of these regulatory sequences and operators within the promoter region specify the promoter architecture.
Genome sequencing and annotation reveal the identity and placement of the TF operators in natural promoters (Collado-Vides et al, 1991; Gralla and Collado-Vides, 1996; Salgado et al, 2006). In these and related works, the distributions of TF operators in Escherichia coli have highlighted trends in the operator positions relative to the polymerase box sequences. For example, it was found that activator operators occur principally around −40, whereas repressor operators were clustered from −60 to +20. These studies proposed that activation is effective only on promoters with low unregulated activity, such as in promoters containing a weak −35 box. The ‘effective repression' of a promoter, defined as the ratio of expression in ‘on' and ‘off' states, was expected to be highest for promoters of strong unregulated activity. These results indicated that repression and activation are most effective at different promoter locations and on different intrinsic promoter strengths.
The potential diversity of promoter architecture and functionality is large when one considers the many known mechanisms by which proteins and DNA interact. Here, we focus on the simplest promoter architectures regulated by multiple TFs and ask what types of regulation functions are possible. Classical descriptions of gene networks have used Boolean logic to describe combinatorial regulation (Kauffman, 1969; Thomas and D'Ari, 1990). However, because the output of a promoter is not a binary function of the concentrations of its regulators (Setty et al, 2003; Atsumi and Little, 2006; Guido et al, 2006; Mayo et al, 2006), a range of non-Boolean logical phenotypes are possible. Recent theoretical descriptions of transcriptional logic (Buchler et al, 2003; Bintu et al, 2005b; Hermsen et al, 2006) have focused on the effects of explicit TF–TF contacts and operator overlap, but it is not known whether such interactions are necessary to generate diverse phenotypes.
To better understand natural promoter function and to improve the design of new promoters for synthetic biology applications (Hasty et al, 2002; Endy, 2005; Sprinzak and Elowitz, 2005), we report a synthetic library-based approach for construction and analysis of modular combinatorial promoters. Here, we varied the placement, affinity, and sequence of known operators (Supplementary Table S1), allowing us to determine the range of functions encoded by the simplest combinatorial promoters. This approach reveals fundamental features of the relationship between promoter architecture and function.
Results
Combinatorial library design and assembly
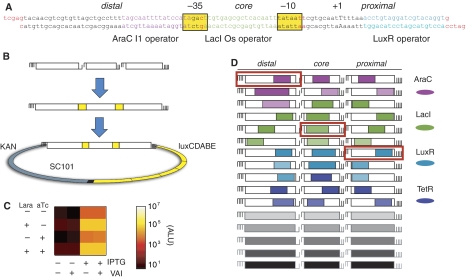
We developed an efficient method for assembling promoters from modular components. The method uses three classes of synthetic duplex DNA units with compatible 5′ cohesive ends. These units correspond to the 45 bp region upstream of the −35 box (distal), the 25-bp region between the −35 and −10 boxes (core), and the 30-bp region downstream of the −10 box (proximal). In this scheme, an arbitrary promoter can be assembled from any combination of proximal, core, and distal units. The internal 5′ overhangs determine each unit's placement in the promoter (Figure 1A). We assayed promoter activity using a bacterial luciferase reporter cassette on a low copy plasmid (Figure 1B and C). Here, we report all promoter activities in terms of arbitrary luminescence units (ALU).
Figure 1.
Random assembly ligation generates a diverse promoter library. Promoters can be assembled out of modular sequence units. (A) The assembled sequence of an example promoter. The 5′ overhangs of each unit are shown in red. The RNA polymerase boxes (−10 and −35) are highlighted in yellow, and the predicted start site of transcription (+1) is capitalized. Operator colors are consistent throughout the figure. (B) Steps in promoter assembly and ligation into the luciferase reporter vector: promoters are assembled by mixed ligations using 1-bp or 2-bp cohesive ends, and then ligated into a luciferase reporter plasmid. (C) Luminescence measurements in 16 inducer conditions (±each of four inducers, as indicated) for the promoter shown in (A). The output levels determine promoter logic. Note that this promoter does not respond to LuxR regulation at the distal region. (D) The 48 unique units used in the library contain operators responsive to the four TFs (indicated by color) in the regions distal, core, and proximal (Sequences in Supplementary Table S1). The promoter fragments corresponding to (A) are boxed in red.
We incorporated operators for two activators and two repressors: The activator AraC (Ogden et al, 1980; Schleif, 2003) regulates arabinose metabolism in E. coli, whereas LuxR activates luminescence genes in Vibrio fischeri (Fuqua et al, 1994). The repressor LacI (Jacob and Monod, 1961; Setty et al, 2003) controls the metabolism of lactose in E. coli, whereas TetR represses the tetracycline resistance genes in transposon Tn10 (Beck et al, 1982; Skerra, 1994). The two activators are active only in the presence of the corresponding inducers L(+)-arabinose (Lara) and oxo-C6-homoserine lactone (VAI), respectively. The repressors TetR and LacI are inactivated by the inducers anhydrotetracycline (aTc) and isopropyl β-D-1-thiogalactopyranoside (IPTG), respectively. Consequently, induction of each factor (AraC, LuxR, LacI, or TetR) is expected to increase a target promoter's activity. These four TFs bind specifically to well-defined operators, are dispensable, and can be induced by small molecules without disrupting normal cellular processes.
For each position (distal, core, and proximal), we designed 5 unregulated and 11 operator-containing units. These sequences varied the affinity, location, and orientation of operators (Figure 1D). The design also allowed for variable −10 and −35 boxes to encourage diverse expression levels. The 16 units of each type were assembled by randomized assembly ligation (Materials and methods) to generate a plasmid library containing approximately 22 000 independent assemblies and providing fivefold coverage of the 163=4096 possible promoters. We transformed the plasmid library into E. coli strain MGZ1X expressing LacI, TetR, AraC, and LuxR. We then sequenced a set of 288 randomly chosen transformants and found 280 correctly assembled promoters (Supplementary Information 1). We determined 217 of these promoters to be unique. Within this set, 47 out of the 48 possible units were represented at least once. Thus, the randomized assembly ligation method produced a diverse set of correctly assembled promoters.
Library functions
We measured the expression of the 288 sequenced transformants in each of 16 combinations of the four chemical inducers (Figure 1C, Materials and methods, Supplementary Information 2). The library showed five decades of variation in promoter activity (Supplementary Figure S1). Promoters of high unregulated activity contained strong −10 and −35 boxes, although the presence of consensus box sequences did not predict unregulated promoter activity (Supplementary Figure S2). Of the 217 unique promoters, 83% produced measurable expression in at least one of the 16 conditions, and 49% changed expression by a factor of 10 or more. Of these 106 clones, 79 were found to respond to a single inducer and 27 responded by more than twofold to two inducers. No promoters were found to respond more than twofold to three or four inducers, or to decrease expression to less than half in the presence of an inducer (anti-induction). All of the dual-input promoters measured increased their activity monotonically in response to the inducer concentrations, both singly and in combination. Overall, the promoter library exhibited a diverse set of behaviors across the 16 conditions.
How does promoter architecture constrain function? For each promoter, we compared the architecture (Supplementary Information 1) with the measured response (Supplementary Information 2). We found no significant regulation without the presence of a corresponding operator (Supplementary Information). The relationship between sequence and phenotype revealed several rules relating promoter architecture to promoter function, which we describe below.
Single-input gates
The simplest promoters termed single-input gates (SIGs) responded to a single inducer (Figures 2 and 3). For these switch-like gates, we defined the regulatory range, r, as the ratio of the induced to uninduced activity. Within this group, activated SIGs showed regulatory ranges up to r=103, whereas the repressed SIGs exhibited higher regulatory ranges up to r=105 (Table I).
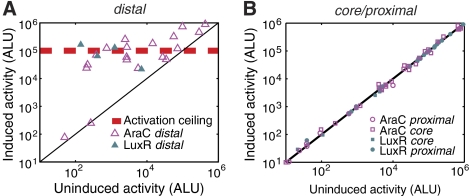
Figure 2.
Activation functions at distal and is attenuated by intrinsic promoter strength. (A) Measurements of promoters activated at distal operators. These promoters respond only to LuxR (solid triangles) or AraC (open triangles) induction. Some promoters fail to respond even though they contain a functional operator (points on the solid line). The activation ceiling (red dashed line) represents the maximal observed activation and does not depend on the unregulated expression level. (B) Promoters containing operators at core (squares) or proximal (circles) do not respond to induction.
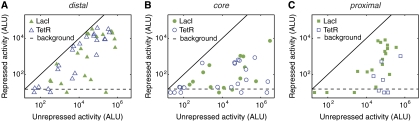
Figure 3.
Repression is effective at all three positions, following the trend core⩾proximal⩾distal. Measurements of repressed single-input promoters. Responses are colored according to the repressor: LacI (filled) or TetR (open). Each promoter contains a single operator located at distal (A), core (B), or proximal (C) positions. Single-input activities are plotted in the induced (unregulated) versus uninduced (repressed) states. In some promoters, operators do not effectively repress the promoter (points located near solid black line). Luciferase detection limits are shown with gray dashed lines.
Table 1.
SIG promoters
| TF | Expression |
Genotypea |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Uninduced (ALU) | Induced (ALU) | Regulatory range (r) | ID | distal | −35 | core | −10 | proximal | |
| TetR | 26±8 | 2.3±0.2 × 106 | 8.9±0.3 × 104 | D18 | con0 | TTGACA | tet1 | GATACT | con1 |
| 14±4 | 1.7±0.1 × 105 | 1.2±0.4 × 104 | B10 | con0 | TTGACA | tet1 | GATACT | con3 | |
| 14±3 | 4.7±0.2 × 105 | 3.3±0.7 × 104 | B19 | ara3 | TTGACA | tet2 | GATACT | tet1 | |
| 23±7 | 9.9±0.8 × 104 | 4.4±1.4 × 103 | A9 | con0 | TTGACA | tet2 | TAGAGT | ara2 | |
| 15±5 | 6.9±1.1 × 104 | 4.7±1.6 × 103 | B22 | con4 | TTGACA | con4 | TAGATT | tet1 | |
| LacI | 12±0 | 3.0±0.2 × 105 | 2.5±0.2 × 104 | B4 | lac1 | TTGACA | lac2 | GATACT | con0 |
| 18±3 | 2.7±0.3 × 104 | 1.5±0.2 × 104 | A81 | lac2 | TTGACT | con1 | GATACT | lux1 | |
| 86±25 | 3.1±0.2 × 105 | 3.6±1.1 × 103 | A38 | lac1 | TTGACA | con0 | TATAAT | lac4 | |
| 273±92 | 7.2±0.2 × 104 | 2.6±0.9 × 102 | A87 | con0 | TTGACA | ara1 | GATACT | lac4 | |
| 58±17 | 3.6±0.3 × 104 | 6.2±1.9 × 102 | A52 | lac1 | TTGACA | ara1 | GATACT | con1 | |
| LuxR | 130±50 | 1.4±0.1 × 105 | 1.1±0.5 × 103 | D49 | lux1 | TTTACT | con2 | GATAAT | lux2 |
| 420±310 | 9.7±2.3 × 104 | 230±180 | D80 | lux3 | TTTACA | lux2 | TATAAT | con3 | |
| AraC | 310±60 | 7.3±0.9 × 104 | 230±50 | A79 | ara2 | TAGACA | ara1 | GATACT | lux2 |
| 250±30 | 2.8±0.6 × 104 | 110±30 | A1 | ara2 | TAGACT | con2 | GATAAT | con1 | |
aThe genotype refers to the three units that make up each promoter and the −10 and −35 polymerase boxes. Here, con, tet, lac, ara, and lux refer to no operator, and TetR, LacI, AraC, and LuxR operators. In each case, the number refers to the operator variant. Full sequences for each unit are available in Supplementary Table S1. Functional operators are highlighted in bold.
Activated SIGs
Activated expression level was independent of unregulated activity (Figure 2A). The best-activated SIGs (highest r) occurred at promoters with low expression in the unregulated state. Activation was ineffective for promoters with unregulated activity above approximately 105 ALU, which is 40-fold lower than the strongest promoter activity measured. This ‘activation ceiling' was the same for both AraC- and LuxR-activated promoters. These results show that activation is limited by the absolute expression level and is most effective on promoters of low intrinsic activity, consistent with previous suggestions (Busby and Ebright, 1994; Gross et al, 1998).
Activation functioned only at the distal position (Figure 2), in accordance with previous studies of AraC and LuxR (Collado-Vides et al, 1991; Egland and Greenberg, 1999). We found neither inducible activation nor inducible repression by LuxR or AraC at core or proximal (Figure 2B). In such promoters, the typical induction response was only 6% for LuxR and 11% for AraC regulation. Some of the strongest activated SIGs (Table I) had additional activator operators at core or proximal sites, along with a functional operator at distal. We found that activator binding to core and proximal did not, on average, strongly affect the maximal promoter activity (Supplementary information, Supplementary Figure S3). These results show that AraC and LuxR have neither positive nor strong negative regulatory effects on gene expression at the core and proximal regions.
Repressed SIGs
In contrast to activation, repression occurred effectively at all three positions (Figure 3). However, we found a clear trend between operator location and repression. Repression was most effective at core (Figure 3B), followed by proximal (Figure 3C), and then distal (Figure 3A). Within this trend, we found that the promoters of low unregulated activity were less sensitive to operator position. This result shows that repression is effective at all three positions, with relative strength following the rule core⩾proximal⩾distal.
As with activation, the expression level in the repressed state was not determined by the unregulated level. Examples of completely repressed expression were observed at every level of unregulated promoter activity (Figure 3). In fact, some repressed SIGs exhibited the highest activities observed (>106 ALU) upon induction (Table I). Within the limits of detection, the effective repression (r) tended to increase with unregulated expression level.
Strikingly, the SIG showing the strongest regulation (r=8.9 × 104, Table I, D18) had only a single TetR operator at the core region. Furthermore, a single repression site at any of the three positions was often enough to repress the promoter below the detection limit (Figure 3). In general, multiple operators were not more effective at repression than single operators. We found nine LacI-regulated and six TetR-regulated SIGs containing multiple repressor operators. Of these, only one LacI-regulated (Table I, A38) and one TetR-regulated (Table I, B19) promoter produced higher regulation than corresponding promoters containing a single operator. These results show that operator position is more important than operator multiplicity for achieving strong regulation with repressors.
Dual-input logic
We next considered dual-input gates as logic functions of their two input inducers. Because of the continuous nature of the output levels in each input state, Boolean logic does not accurately represent the space of possible functions. For example, in a recent study, the natural lac promoter increased activity by a factor of 3.6 when induced by cAMP alone, by a factor of 7.1 when induced by IPTG alone, and by a factor of 14 when induced by both simultaneously (Setty et al, 2003). This intermediate behavior could be described as either AND-like or OR-like, depending on the activity threshold chosen.
To describe such ‘intermediate logic' phenotypes, we introduced a three-dimensional parameterization for the space of promoter functions. In this scheme, we represented the promoter functions with three numerical parameters that quantify dynamic range, logic type, and asymmetry of inputs (Materials and methods). As before, r is the ratio of the maximum to minimum promoter activity. The parameter l quantifies the logical behavior of the promoter: from pure OR (l=0) to pure AND logic (l=1). Finally, the parameter a quantifies the asymmetry of the gate with respect to its two inducers. At a=0, the gate responds identically to either inducer, whereas at a=1, the promoter responds to one input only (pure SIG). These parameters span the full range of observed phenotypes and have intuitive interpretations. They also represent relative promoter activities rather than absolute levels, making them less sensitive to the choice of reporter, growth media, or other experimental conditions. Therefore, they form an ideal quantitative representation for the phenotypic behavior of these promoters.
Within this logic-symmetry space, the positive monotonic response of promoters to their inputs restricts promoter logic to the triangular region shown in Figure 4. The corners of this region include three Boolean logic functions: the switch-like SIG (l=0.5, a=1), along with the canonical binary gates AND (l=1, a=0) and OR (l=0, a=0). The symmetric SLOPE gate (l=0.5, a=0) exhibits logic intermediate between AND and OR. The asymmetric asym-AND (l=0.75, a=0.50), asym-OR (l=0.25, a=0.50), and asym-SLOPE (l=0.50, a=0.50) gates describe idealized logic functions intermediate between SIG and AND, SIG and OR, and SIG and SLOPE, respectively (Figure 4A). This representation provides qualitative categories for the different types of logic displayed by monotonic dual-input promoters.
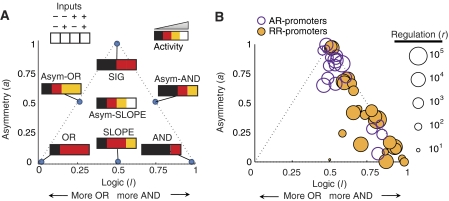
Figure 4.
Dual-input gates exhibit diverse functions in logic-symmetry space. Promoter response phenotypes can be represented by their asymmetry, a (y-axis), logic type, l (x-axis), and regulatory range, r. (A) Diagram showing the space of allowed logical phenotypes, with the locations of ideal logic gates indicated. The SIG responds completely to one inducer and not at all to the other. The SLOPE gate represents an intermediate logical function between AND and OR, whereas the asymmetric gates are intermediate between SIG and the corresponding symmetric gate. Intermediate logical behavior is represented between these ideal locations. The logic-symmetry parameterization is defined in the Materials and methods. Points outside of the dashed triangle are not accessible if promoters respond monotonically to each input. (B) The logical phenotypes of 50 dual-input promoters (r>3). AR promoters are shown as purple circles, RR promoters are shown as gold disks. The diameter of each disk is proportional to the logarithm of its regulatory range, r.
We identified 50 dual-input gates (Materials and methods). Each defined a point (r, a, l) in the logical phenotype space (Figure 4B), revealing a range of functional behaviors. Asym-AND and SIG-like gates exhibited strong regulation up to r=105. The AND and asym-SLOPE gates were regulated up to r=104, whereas the SLOPE gates were regulated up to r=103. Notably, we found no gates exhibiting strong OR or asym-OR logic functions. However, one class of dual-input promoters (discussed below) exhibited asym-SLOPE logic approaching an asym-OR response (l<0.50). Thus, we observed a wide distribution of promoter logic types.
The library contained two classes of dual-input gates. The repressor–repressor (RR) promoters contained operators for the repressors LacI and TetR, whereas the activator–repressor (AR) promoters responded to the activator AraC and one of the repressors. Due to the relative scarcity of LuxR-activated promoters, we did not find LuxR-regulated AR promoters in the characterized promoter set (Figure 2A). These two classes of dual-input gates exhibited differing but overlapping distributions of logical phenotypes.
Comparison of AR and RR promoter phenotypes (Figure 4B) revealed that each has a preference for different logical categories, although both produced strong asym-AND gates. The RR promoters produced the strongest symmetric (AND and SLOPE) gates, whereas the AR promoters generated the strongest asym-SLOPE gates. This shows that RR promoters produced both symmetric and asymmetric logic, while AR promoters produced only asymmetric logic.
Mathematical model of repressor interaction
To better understand the variety of symmetric and asymmetric logic observed for the RR promoter class, we employed a simple model of promoter activity in the presence of two repressors (Materials and methods). In this model, c1, c2, and ω represent the strength of repression at the stronger operator, the weaker operator, and the repressor–repressor interaction, respectively (Bintu et al, 2005b). When the repressors do not interact with each other, ω=1, whereas for exclusive interactions (only one repressor can bind at a time), ω=0. Cooperative interactions would correspond to ω>1.
The logic parameter l was tightly coupled to the model interaction parameter ω (Materials and methods). A plot of a and l as parameterized functions of the microscopic model parameters (Supplementary Figure S4) showed that RR promoters with ω ranging from 0 (exclusive interaction) to 1 (independent interaction) can produce any logic function in the right half (l⩾0.5) of the phenotype space triangle: SIG, AND, SLOPE, asym-AND, and asym-SLOPE. In particular, exclusive interaction (ω=0) approached pure AND logic (l=1), whereas independent interaction (ω=1) always resulted in SLOPE-like logic (l=0.5). Conversely, we found that an asym-OR gate would require extremely high cooperative interaction (ω=100), whereas an ideal OR gate would require infinite cooperativity. Therefore, the range of logic functions displayed by the library RR promoters (Figure 4B) falls within the spectrum of noncooperative interactions (1⩾ω⩾0). This model demonstrates that a variety of logic functions can be achieved without explicit protein–protein cooperativity.
RR promoters
Dual repression can be either symmetric or asymmetric (Figure 4B), with either repressor dominant (Figure 5A). As with the SIGs, even the strongest RR promoters could be fully repressed, exhibiting effective repression up to r=105. RR promoter logic was always AND-like or SLOPE-like (0.5⩽l⩽1.0), indicating that there were no instances of strong cooperative interaction between the repressors (ω⩽1). In three cases, mutation of a repressor operator resulted in almost completely asymmetric (a=1) SIG logic (Figure 4B, top of triangle). In other cases, the repression was more balanced (a<0.25), producing symmetric AND and SLOPE responses. Thus, RR promoters displayed a large range of dual-input regulatory logic including AND, SLOPE, asym-SLOPE, and asym-AND gates.
Figure 5.
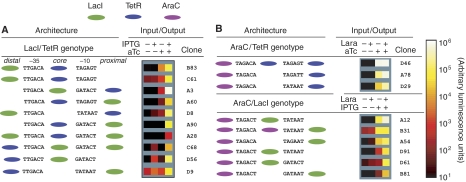
Combinatorial promoter architecture reveals rules for programming gene expression. The architecture and function of several dual-input promoters. The architecture of each promoter (colored according to Figure 1) is shown with its functional operators and −10 and −35 boxes. (A) RR promoters respond to both LacI and TetR. The fourth induction column (+ IPTG, +aTc) corresponds to the unregulated state. (B) AR promoters respond to AraC and one of the two repressors, as indicated. Here, the third column (+IPTG/aTc, −Lara) corresponds to the unregulated state.
In principle, the logic phenotype displayed by a promoter could depend on the inducer concentrations used. Therefore, we chose three RR promoters (Figure 5A, clones A3, D8, and D9), and measured their responses to 16 combinations of inducer concentrations (Supplementary Information). As expected, all three promoters increased their activity monotonically with increasing concentrations of each inducer. As shown in Supplementary Figure S5, inducer concentrations primarily affected r and a, whereas the logic parameter l was less dependent (Supplementary Information). The most AND-like gate (A3) had the highest variation in logic (l=0.46 to l=0.86), whereas the most SLOPE-like (D9) exhibited the narrowest range (l=0.48 to l=0.53). These results imply that r and a depend strongly on input concentration; whereas for l, independent (SLOPE) logic is more robust than exclusive regulation (AND).
The repressor operator location trend core⩾proximal⩾distal explains the combinatorial promoter behaviors shown in Figure 5A. For RR promoters, the position of the operators determined whether LacI or TetR was dominant. We found only one clear exception to this trend (Figure 5A, clone A3), where TetR acting at proximal slightly dominates LacI acting at core. Symmetric repression occurred for several architectures, such as with a TetR at core and two LacI operators, one at distal and the other at proximal (Figure 5A, A28). In all other asymmetric cases core dominated proximal and distal, while proximal dominated distal. RR promoter architectures with operators at proximal and distal produced the largest range of logic behaviors including AND, SLOPE, asym-AND, and asym-SLOPE. RR promoters with operators at the core and proximal positions produced only AND and asym-AND logic. Of the seven RR promoters exhibiting strong AND-like logic (l>0.8), five had operators at core and proximal. Finally, RR promoter architectures with operators at core and distal produced the most asymmetric logic functions (e.g. Figure 5A, B83); the repressor acting at core was always strongly dominant. These results show that repressor dominance in combinatorial promoters follows the trend core⩾proximal⩾distal, and that close operator proximity is consistent with AND-like logic.
AR promoters
Among AR promoters (Figure 5B), repression always dominated activation (0.06⩽a⩽0.99). The AR promoters were regulated by AraC, in combination with LacI or TetR and exhibited regulation up to r=104. In all cases, the activator functioned from the distal region, whereas the repressor functioned at core or proximal. We found one AR promoter that approached symmetric response (r=3272, a=0.06, l=0.81, Figure 5B, D61). The three most AND-like (l>0.8) promoters of this class had the repressor operator at the core. The most OR-like (smallest l) promoter exhibited asym-SLOPE logic (r=9112, a=0.65, l=0.46, Figure 5B, A54), with the repressor operator at proximal. Therefore, we found AR promoters are well represented by asym-AND when the repressor acts as core and asym-SLOPE when the repressor acts at proximal.
The AR promoters also confirmed our previous result relating activation to intrinsic promoter activity: the higher the unregulated activity of an AR promoter (+IPTG/aTc, −Lara), the smaller the change upon activator induction (compare the last two columns in Figure 5B). When the unregulated activity exceeded the activation ceiling, the AR promoter did not respond to AraC induction at all, resulting in SIG-like behavior (e.g. Figure 5B, D46). This result indicates that AR promoters will depend on both inputs only when the unregulated promoter activity is below the activation ceiling.
Discussion
Combinatorial synthesis of synthetic promoters, as described here, permits systematic analysis of promoter architecture and rapid identification of promoters that implement specific functions. The spectrum of promoter functions observed in this library highlights several heuristic rules for promoter design:
Limits of regulation. Gene expression can be regulated over five orders of magnitude. Regulated promoter activity is independent of unregulated activity. As a result, effective repression tends to increase with unregulated activity, whereas activation tends to decrease. Activation is limited by an absolute level of expression, at around 2.5% the level of the strongest unregulated promoter activities.
Repressor operator location. The effectiveness of repression depends on the operator location with core⩾proximal⩾distal. Dual-repression may be symmetric or asymmetric, with the dominant repressor predicted by operator locations.
One is enough. Full repression is possible with a single operator between −60 and +20 at high repressor concentrations. Activators function only upstream of −35 (distal), and have little positive or negative effect downstream at core or proximal.
Repression dominates activation, producing asymmetric logic.
Operator proximity. Independent regulators generate SLOPE-like logic. Operator proximity increases competitive interactions, making the logic more AND-like.
For both activation and repression, the activity of the promoter in the regulated (activated/repressed) state is not determined by the activity in the unregulated state (Rule 1). Intuitively, activation has higher r when the unregulated activity is low, and repression has higher r when the unregulated activity is high. Furthermore, as predicted by recent theoretical work (Bintu et al, 2005a), repression is able to achieve extremely high levels of regulation (r⩽105), whereas activated regulation is moderately strong (r⩽103). These limits apply to both SIGs (Figures 2 and 3) and dual-input promoters (Figure 5). AR promoters are a special case and exhibit a trade-off: increasing the unregulated activity increases the regulatory range (r) at the expense of greater asymmetry (a). For example, compare the first and last promoter in Figure 5B.
Rules 2 and 3 summarize the operator position and multiplicity effects for both activators and repressors. The repression trend (Rule 2) has been previously reported for promoters regulated by LacI (Lanzer and Bujard, 1988; Elledge and Davis, 1989). The authors of the first paper proposed a mechanistic model involving two competing effects: core and proximal sites more effectively block polymerase binding, whereas core and distal sites bind to repressor more rapidly (are more accessible) as the polymerase initiation complex clears the −10 and −35 boxes. We confirmed the operator location trend for SIGs regulated by LacI and TetR alone and found that this heuristic also holds for RR promoters of both repressors. Of course, differences in operator affinity, repressor concentration, and repressor structure can overcome these rules.
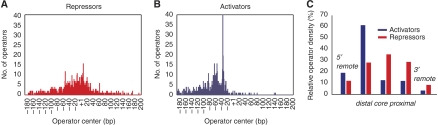
We compared Rules 2 and 3 with the distribution of known E. coli operators compiled from 1102 natural promoters in the database RegulonDB (Salgado et al, 2006; Figure 6). In agreement with analysis made on earlier versions of the database (Collado-Vides et al, 1991; Gralla and Collado-Vides, 1996), we found that activator operators are most common in the distal region (Figure 6A), whereas repressor operators cluster around all three promoter regions (Figure 6B). Figure 6C shows the operator density of the 554 promoters that are recognized by the polymerase subunit σ70. The small regulatory effect observed for activator operators in the core and proximal regions (Rule 3) appears consistent with the general scarcity of natural activator sites in these regions. Similarly, the density of repressor operators found in σ70 promoters is significantly enriched for core sites over distal and proximal locations, consistent with the repressor operator location trend (Rule 2).
Figure 6.
The distribution of operator locations in natural promoters reflects functional trends of synthetic promoters. Operator locations are as annotated in RegulonDB 5.0 (Salgado et al, 2006). Distributions of repressor (A) and activator (B) operators as found in 1102 E. coli promoters. The number of operators centered at each position relative to the start site of transcription (+1) is plotted. (C) The density of operators found in 554 σ70 promoters is broken down into three promoter regions, distal, core, and proximal, as well as regions upstream (5′ remote) and downstream (3′ remote) of the promoter. The density is shown as the fraction of sites in each position weighted by the relative size (bp) of each region.
The sufficiency of one operator for repressing promoter activity up to five orders of magnitude (Rule 3) raises the classic question of why natural promoters are so often regulated by redundant operators (Collado-Vides et al, 1991). Our study used high concentrations of repressors in the range of 2–4 μM (Lutz and Bujard, 1997), paired with strong operators (Supplementary Table S1). At lower repressor concentrations and operator affinities, the presence of multiple binding sites can increase the effective repression r through looping (Vilar and Leibler, 2003; Becker et al, 2005), cooperativity (Oehler et al, 1994; Ptashne, 2004; Rosenfeld et al, 2005), or even without explicit TF–TF interactions (Bintu et al, 2005a). These effects can also increase the steepness of response to repressor concentration (Ptashne, 2004), or engender exceptions to the dominance of repression (Rule 4). Finally, the presence of multiple operators might increase the mutational plasticity of promoter functions (Mayo et al, 2006).
Rule 5 provides insight for both AR and RR promoters: operators at the neighboring sites will tend to generate more AND-like logic (higher l) than non-neighboring sites (i.e. distal and proximal). In AR promoters, repression at core produces more AND-like logic than at proximal. This effect can be understood intuitively for RR promoters: if operators are closely spaced, binding of one repressor can inhibit the binding of the other. Removing one repressor has two conflicting effects: it increases expression due to its reduced occupancy, but it simultaneously decreases expression by allowing binding of the other repressor. This makes the overall logic more AND-like. In terms of the mathematical model, AND-like (l>0.8) RR promoters correspond to strong balanced repression (c1≈c2≫1) and exclusive interaction (ω≈0).
The library described here represents a starting point for systematic investigation of the functional repertoire of prokaryotic promoters. These simple promoters cannot include all the complex effects found in natural promoters, including those dependent on DNA bending or specific protein–protein interactions. Nevertheless, they provide a view of what is possible with the simplest genetic elements and interactions. Within this context, the heuristics described above allow the design of particular promoter functions controlled by arbitrary TF regulators. The assembly method allows for construction of any specific promoter. Other promoter architectures could be generated with this method to provide more diverse logic phenotypes, or to explore regulatory DNA in eukaryotic organisms (Ligr et al, 2006). For example, the lac promoter architecture, regulated by a distal activator and multiple repressor operators (including upstream sites), can exhibit phenotypes not found in our library, such as asym-OR (Mayo et al, 2006). In another case, a synthetic activator–activator (AA) promoter has been constructed, which exhibits near-symmetric SLOPE logic (Joung et al, 1994). Tandem promoters are expected to generate additive logic functions more closely representing OR logic, and in fact, many natural promoters are found in tandem repeats (Collado-Vides et al, 1991). If our heuristic rules apply to natural combinatorial promoters, we may begin to elucidate complicated functions by inspection of these non-coding DNA sequences. In this regard, effective parameterizations of logic, such as the one shown in Figure 4, can provide a more intuitive understanding of the computations performed by promoters.
Materials and methods
Reagents
All inducers and chemicals were purchased from Sigma. Concentrations (unless otherwise stated) were 50 μg/ml kanamycin, 100 μg/ml ampicillin, 500 μM IPTG, 100 ng/ml anhydrotetracycline (aTc), 0.1% L(+)-arabinose (Lara), and 1 μM oxo-C6-homoserine lactone (VAI). LB growth media (Lennox) was used for all experiments. All ligation reactions were carried out with 1.25 U of T4 DNA ligase (Invitrogen) and 0.1 mg/ml BSA (Invitrogen) in 20 μl of T4 ligase buffer (Invitrogen) at 4°C.
Randomized assembly ligation
Promoters were constructed by total synthesis and ligation. Each promoter was constructed from three duplex DNA fragments comprising the distal, core, and proximal regions. An overhanging phosphorylated G on the downstream 5′ end of distal is compatible with a phosphorylated overhanging C on the upstream 5′ end of core. Likewise, an overhanging phosphorylated AA on the downstream 5′ end of core is compatible with an overhanging phosphorylated TT on the 5′ upstream end of proximal. The terminal ends of the fully assembled promoters had mutually incompatible XhoI and BamHI 4 bp 5′ overhangs, which remained unphosphorylated. A total of 48 duplex units (Supplementary Table S1) were annealed out of 96 PAGE-purified synthetic DNA oligonucleotides (University of Calgary DNA synthesis and sequencing center) at 1 μM in T4 ligase buffer. All 48 duplex units were mixed together in equal 50 nM proportions and ligated for 1 week, then cloned into bacterial luciferase reporter plasmid pCS26 (Bjarnason et al, 2003). We purified the plasmids using the Qiagen Plasmid Midi kit and transformed the library into strain MGZ1X (reference MG1655 (Riley et al, 2006) containing the native ara operon, the LacI- and TetR-overexpressing Z1 cassette (Lutz and Bujard, 1997), and the medium-copy plasmid pCD136 which constitutively expresses LuxR). We picked 10 000 clones and chose 288 randomly for sequencing (Bjarnason et al, 2003) and functional characterization.
Luminescence measurements
The library was assayed in 16 inducer conditions corresponding to all saturating combinations of the four inducers: VAI, IPTG, Lara, and aTc. Cells were grown in 96-well plates to stationary phase (16–22 h at 37°C) and inoculated into triplicate 96-well plates containing LB media, antibiotics, and each inducer combination. These were grown at 25°C for 18 h in the dark. Luminescence measurements were obtained using a Tecan Safire plate reader (100 ms integration, default settings). To determine the background, we took the median measurements of nonfunctioning clones in each condition. All data reported are the median of triplicate measurements.
To assess the luminescent crosstalk between neighboring wells, we inoculated a constitutively bright clone into every other row and column of a 96-well plate (total 24 wells) and measured it continuously during growth over 18 h. These data were used to compute the horizontal/vertical (j1) and diagonal (j2) neighbor crosstalk. We assumed (linear) crosstalk of the form O=AX, where O is the observed data, A the actual luminescence of each well, and X the crosstalk matrix. We computed A=OX−1 for combinations (j1, j2) and then took the total variance of all empty wells as a metric. This metric reached a minimum of 0.017% horizontal/vertical and 0.002% diagonal crosstalk. This was a very small effect compared to other sources of error (below), and only resulted in an appreciable difference for wells neighboring the very brightest clones (∼106 ALU). The vector background level (∼10 ALU) was subtracted from all data points. We set each datum to a minimum level of 10, corresponding to 1 count/100 ms.
To assess the plate-to-plate variation, we calculated the standard error between triplicates and divided by the mean. We found an average replicate error of 24%. To assess day-to-day error, we measured one set of 96 clones on two consecutive days and computed standard relative errors by a linear fit of the second day's data to the first (44%). Similarly, we computed the well-to-well error on the same plate by identifying clones with the same sequence genotype and doing a linear fit between them (54%). Together these data provide an upper limit of ∼50% on repeatability.
Promoter function analysis
To calculate the expression levels for dual-input promoters (or SIGs), we first identified the two (one) primary inducers of each promoter. We then averaged the luminescence data over the four (eight) background conditions. Standard errors were computed from these values, and the median of the triplicate measures gave the four (two) expression levels of the gate. We then computed the regulatory ratio r, defined as the maximum expression level divided by the minimum. The error in regulation (Table I) was computed from the relative errors for each state. For SIGs with expression levels b1 (off) and b2 (on), the error in r is
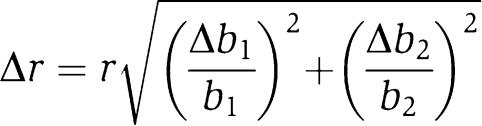 |
We identified SIGs and dual-input promoters from their sequences (Supplementary Information 1). Functional activator operators were found at distal and functional repressor operators occurred at all three positions. With one exception (discussed in Supplementary Information), significant ( × 2) regulation by a TF occurred only with one or more corresponding operators in the promoter sequence. The presence of an operator did not always guarantee regulation: nonfunctioning SIGs lie on the diagonal lines of Figures 2 and 3, and dual-input promoters responding to only one input occur at the apex (a≈1) of the triangle in Figure 4B.
Logic-symmetry space
In addition to the regulation r, the two-input gates displayed a variety of relative expression levels. For the dual-input promoters, we defined four measured response values (b1, b2, b3, b4) such that b4⩾b3⩾b2⩾b1. As repression always dominated activation, for AR promoters b2 corresponded to the activator-induced state and b3 corresponded to the repressor-induced state. Similarly, for RR promoters, b2 corresponded to the expression level when the weaker repressor is induced and b3 to the induction of the stronger. To represent the range of logical functionality, we defined the three phenotypic parameters (r, a, l) in terms of these response values:
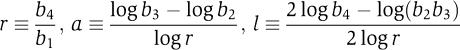 |
Specifically, l quantifies the logic type ranging from a perfect AND (b3=b2=b1 → l=1) to a perfect OR (b3=b2=b4 → l=0). The parameter a quantifies the asymmetry with respect to the two inputs, ranging from perfectly symmetric (b2=b3 → a=0) to the completely asymmetric SIG (b3=b4 and b2=b1 → a=1).
SLOPE theorem: separation of variables in combinatorial gene regulation
Consider a dual-input promoter regulated by two TFs: X and Y (we use x and y to represent their respective activities). If these TFs regulate the promoter independently with single-input functions s(x) and t(y), the variables of the regulation function p(x,y) separate: p(x,y)=s(x)t(y). Suppose (without loss of generality) that regulator X is dominant. Then the four logical output states of the promoter are:
The arrows signify the high (↑) and low (↓) states of the promoter with respect to each input (e.g. induced and uninduced, respectively). The logic parameters of the promoter are then, by definition
Considering the logic parameter l, the separation of variables requires that
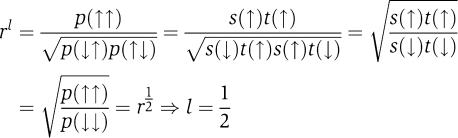 |
Therefore, separation of variables—regardless of the TF regulation functions—implies that the promoter logic is always SLOPE or asym-SLOPE (or in the case that one of the regulators is nonfunctional, SIG). The converse is not generally true, but it does hold for the model of dual repression discussed below.
Model of RR promoter logic
We employed a previously defined model of RR promoter activity under dual repression (Bintu et al, 2005b):
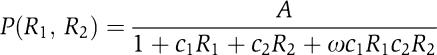 |
The maximal promoter activity is A and the normalized repressor concentrations (R1, R2) range from 0 to 1. Here, c1 and c2 represent the effectiveness of each repressor at excluding polymerase from the promoter. The term ω represents interactions between repressors: ω<1 corresponds to competitive binding, ω=0 represents exclusive binding, and ω>1 represents cooperative binding. When ω=1, the repressors are said to act independently.
We solved for the three logic-symmetry parameters (r, a, l) in terms of the three microscopic parameters (c1, c2, ω):
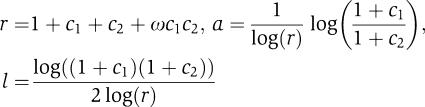 |
By the SLOPE theorem, independent interaction (ω=1) produces SLOPE-like logic (l=0.5). The converse is also true here: when l=0.5, RR promoters (c1⩾c2>0) are regulated by the two repressors independently (ω=1):
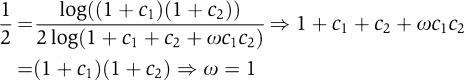 |
For symmetric RR promoters (c=c1=c2 → a=0), the independently interacting RR promoter is an ideal SLOPE gate (a=0, l=0.5). When the interaction is symmetric but dependent (ω≠1), the logic l is described by
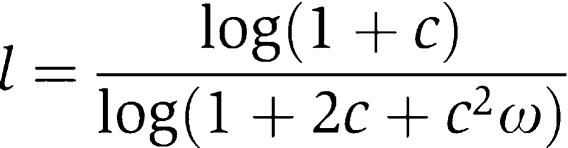 |
For exclusive interaction (ω=0), the logic depends only on the operator strength c. As c grows large, the logic approaches pure AND (l=1):
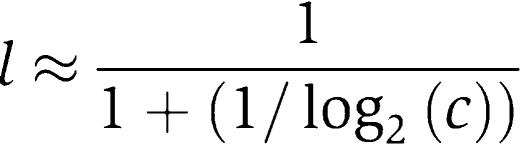 |
In the opposite extreme, pure OR logic (l=0) is only approached in the limit logc ω→∞:
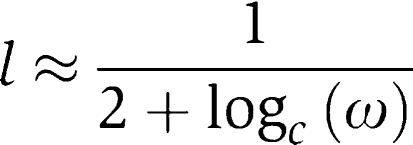 |
RegulonDB analysis
Following prior analysis of TF binding sites (Collado-Vides et al, 1991), we examined 1102 E. coli regulatory promoter sequences from RegulonDB 5.0 (Salgado et al, 2006). Operator binding sites for activators and repressors in each promoter were identified. The TF operators annotated as ‘dual' were removed from this list. For each operator, we determined the middle of the annotated binding sequence; calculated the distance to the annotated transcription start, and calculated the number of repressor and activator operators centered at each base pair in the region (∼400 bp total). These distributions were plotted as histograms for activators and repressors (Figure 6A and B). We also calculated the distribution of operators for 554 promoters recognized by σ70 (Figure 6C). In this histogram, the relative fraction at each region was weighted by its length in bp. This weighting was necessary to observe the enrichment of repressor operator density in the core region.
Supplementary Material
Supplementary Information 1
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Table S1
Supplementary Information 2
Supplementary Information
Acknowledgments
We thank Aaron White for help with library design and construction, Mercedes Paulino for help preparing figures, and Carla Davidson for plasmid pCD136. Avidgor Eldar contributed to the logic-symmetry space formalism. T Irie, D Morris, P Sternberg, E Winfree, G Anderson, J Locke, H Garcia, R Kishony, U Alon, and C Dalal provided helpful discussions. We thank W Kim, C Vizcarra, G Seelig, and J Kim for technical assistance. RSC was partially supported by the National Physical Science Consortium and Sandia National Laboratory. MGS is supported as an Alberta Heritage Foundation for Medical Research (AHFMR) Scientist and Canada Research Chair in Microbial Gene Expression. This work was supported by NIH (grants R01GM079771 to MBE and 5P50 GM068763 to the Center for Modular Biology), HFSP, the Packard Foundation, and the Caltech Center for Biological Circuit Design.
References
- Atkinson MR, Savageau MA, Myers JT, Ninfa AJ (2003) Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 113: 597–607 [DOI] [PubMed] [Google Scholar]
- Atsumi S, Little JW (2006) A synthetic phage lambda regulatory circuit. Proc Natl Acad Sci USA 103: 19045–19050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R (2005) A synthetic multicellular system for programmed pattern formation. Nature 434: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R (2004) Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci USA 101: 6355–6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CF, Mutzel R, Barbe J, Muller W (1982) A multifunctional gene (tetR) controls Tn10-encoded tetracycline resistance. J Bacteriol 150: 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker NA, Kahn JD, Maher LJ III (2005) Bacterial repression loops require enhanced DNA flexibility. J Mol Biol 349: 716–730 [DOI] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Kuhlman T, Phillips R (2005a) Transcriptional regulation by the numbers: applications. Curr Opin Genet Dev 15: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R (2005b) Transcriptional regulation by the numbers: models. Curr Opin Genet Dev 15: 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnason J, Southward CM, Surette MG (2003) Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J Bacteriol 185: 4973–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning DF, Busby SJ (2004) The regulation of bacterial transcription initiation. Nat Rev Microbiol 2: 57–65 [DOI] [PubMed] [Google Scholar]
- Buchler N, Gerland U, Hwa T (2003) On schemes of combinatorial transcription logic. Proc Natl Acad Sci USA 100: 5136–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby S, Ebright RH (1994) Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79: 743–746 [DOI] [PubMed] [Google Scholar]
- Chan B, Busby S (1989) Recognition of nucleotide sequences at the Escherichia coli galactose operon P1 promoter by RNA polymerase. Gene 84: 227–236 [DOI] [PubMed] [Google Scholar]
- Collado-Vides J, Magasanik B, Gralla JD (1991) Control site location and transcriptional regulation in Escherichia coli. Microbiol Mol Biol Rev 55: 371–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egland KA, Greenberg EP (1999) Quorum sensing in Vibrio fischeri: elements of the luxl promoter. Mol Microbiol 31: 1197–1204 [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Davis RW (1989) Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev 3: 185–197 [DOI] [PubMed] [Google Scholar]
- Endy D (2005) Foundations for engineering biology. Nature 438: 449–453 [DOI] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176: 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla JD, Collado-Vides J (1996) Organization and function of transcription regulatory elements. In Escherichia coli and Salmonella: Cellular and Molecular Biology, Neidhardt FC, Curtiss R (eds), 2nd edn, pp 2 v. (xx, 2822 pp) Washington, DC: ASM Press [Google Scholar]
- Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B (1998) The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb Symp Quant Biol 63: 141–155 [DOI] [PubMed] [Google Scholar]
- Guido NJ, Wang X, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC, Collins JJ (2006) A bottom-up approach to gene regulation. Nature 439: 856–860 [DOI] [PubMed] [Google Scholar]
- Hasty J, McMillen D, Collins JJ (2002) Engineered gene circuits. Nature 420: 224–230 [DOI] [PubMed] [Google Scholar]
- Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL (2006) rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell 125: 1069–1082 [DOI] [PubMed] [Google Scholar]
- Hawley DK, McClure WR (1983) Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res 11: 2237–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsen R, Tans S, Wolde PR (2006) Transcriptional regulation by competing transcription factor modules. PLoS Comput Biol 2: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3: 318–356 [DOI] [PubMed] [Google Scholar]
- Joung JK, Koepp DM, Hochschild A (1994) Synergistic activation of transcription by bacteriophage lambda cI protein and E. coli cAMP receptor protein. Science 265: 1863–1866 [DOI] [PubMed] [Google Scholar]
- Kammerer W, Deuschle U, Gentz R, Bujard H (1986) Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in late steps of the overall process. EMBO J 5: 2995–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman S (1969) Homeostasis and differentiation in random genetic control networks. Nature 224: 177–178 [DOI] [PubMed] [Google Scholar]
- Lanzer M, Bujard H (1988) Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci USA 85: 8973–8977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligr M, Siddharthan R, Cross FR, Siggia ED (2006) Gene expression from random libraries of yeast promoters. Genetics 172: 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U (2003) Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA 100: 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo AE, Setty Y, Shavit S, Zaslaver A, Alon U (2006) Plasticity of the cis-regulatory input function of a gene. PLoS Biol 4: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehler S, Amouyal M, Kolkhof P, von Wilcken-Bergmann B, Muller-Hill B (1994) Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J 13: 3348–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden S, Haggerty D, Stoner CM, Kolodrubetz D, Schleif R (1980) The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci USA 77: 3346–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M (2004) A Genetic Switch: Phage Lambda Revisited, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Ptashne M (2005) Regulation of transcription: from lambda to eukaryotes. Trends Biochem Sci 30: 275–279 [DOI] [PubMed] [Google Scholar]
- Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, Mori H, Perna NT, Plunkett G III, Rudd KE, Serres MH, Thomas GH, Thomson NR, Wishart D, Wanner BL (2006) Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res 34: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB (2005) Gene regulation at the single-cell level. Science 307: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL (1993) A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262: 1407–1413 [DOI] [PubMed] [Google Scholar]
- Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, Segura-Salazar J, Martinez-Antonio A, Collado-Vides J (2006) RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res 34: D394–D397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R (2003) AraC protein: a love–hate relationship. BioEssays 25: 274–282 [DOI] [PubMed] [Google Scholar]
- Setty Y, Mayo AE, Surette MG, Alon U (2003) Detailed map of a cis-regulatory input function. Proc Natl Acad Sci USA 100: 7702–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerra A (1994) Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene 151: 131–135 [DOI] [PubMed] [Google Scholar]
- Sprinzak D, Elowitz MB (2005) Reconstruction of genetic circuits. Nature 438: 443–448 [DOI] [PubMed] [Google Scholar]
- Thomas R, D'Ari R (1990) Biological Feedback. Boca Raton: CRC Press [Google Scholar]
- Vilar JM, Leibler S (2003) DNA looping and physical constraints on transcription regulation. J Mol Biol 331: 981–989 [DOI] [PubMed] [Google Scholar]
- Zubay G, Schwartz D, Beckwith J (1970) Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc Natl Acad Sci USA 66: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information 1
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Table S1
Supplementary Information 2
Supplementary Information