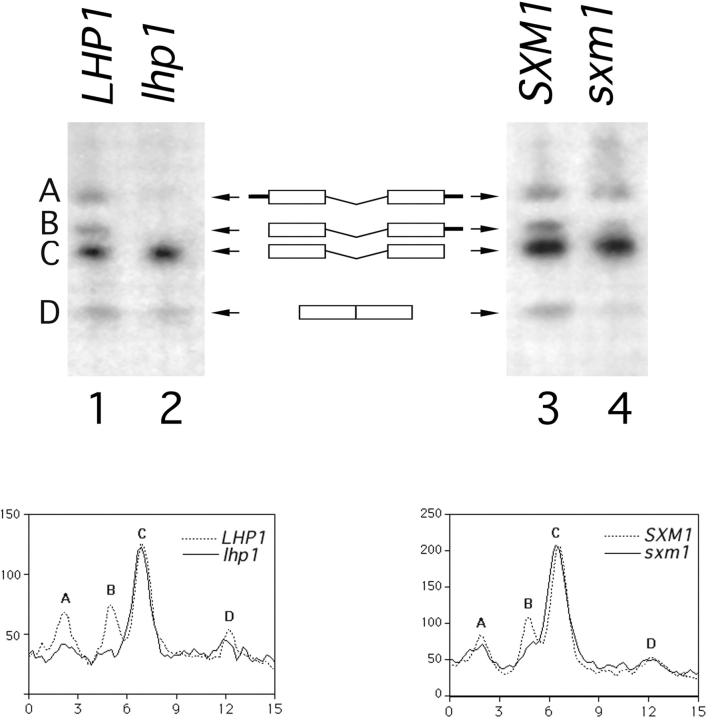
Figure 5.
3′ processing of pre-tRNA Ser CGA Ser CGA in LHP1- and SXM1-deletion strains. A schematic of the intron-containing pre-tRNAs, species A, B, and C and of mature tRNA, species D, is included for reference. Exons are represented by boxes, 5′ and 3′ extensions are represented by black bars, and the central intron is shown. A Northern blot of total RNA was probed for intermediates A, B, and C using an oligonucleotide from the intron, and for mature tRNA using a probe that spans the properly spliced exons. Lane 1, wild-type cells (LHP1); lane 2, lhp1 cells; lane 3, Lhp1-PrA cells, wild-type for SXM1 (SXM1); and lane 4, Lhp1-PrA/sxm1 cells (sxm1). The blot was quantified using a PhosphorImager with ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The graphs indicate radioactivity relative to distance, and peaks corresponding to intermediates are labeled. To account for minor loading discrepancies, the data in the graphs have been normalized to an internal control, precursor C, which was shown previously (Yoo and Wolin, 1997) to be independent of LHP1 genotype.