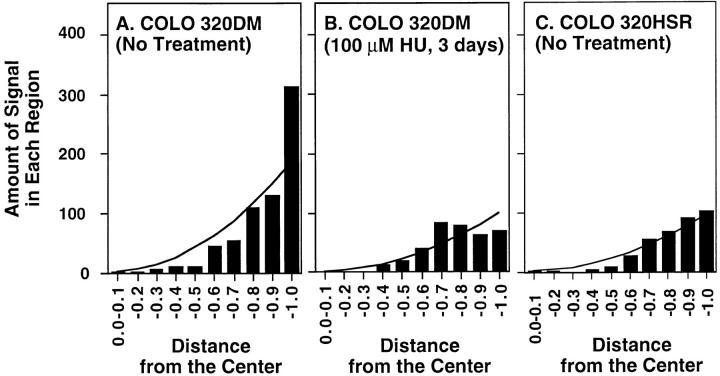
Figure 5.
Quantitative analysis of the nuclear positions of DMs and chromosomally amplified sequences. The nuclei from a rapidly growing culture of COLO 320DM cells (A), COLO 320DM treated with 100 μM HU for 3 d (B), and rapidly growing COLO 320 cells with chromosomally amplified c-myc sequences (COLO 320HSR) (C) were hybridized with a probe for the c-myc amplicon. Sections intercepting the center of each nucleus were obtained as in Fig. 4. For each section, the position and intensity of each hybridized signal was measured. Each analysis represents measurements on 100 randomly chosen nuclei. The abscissa depicts the fractional distance from the nuclear center (0, center; 1, periphery), and the ordinate is the number of signals detected at each position in 100 nuclei. The theoretical random distribution curves based on signals per nuclear vol at each position are shown in each graph as explained in greater detail in Materials and Methods.