Abstract
Chromosome arrangement in spread nuclei of the budding yeast, Saccharomyces cerevisiae was studied by fluorescence in situ hybridization with probes to centromeres and telomeric chromosome regions. We found that during interphase centromeres are tightly clustered in a peripheral region of the nucleus, whereas telomeres tend to occupy the area outside the centromeric domain. In vigorously growing cultures, centromere clustering occurred in ∼90% of cells and it appeared to be maintained throughout interphase. It was reduced when cells were kept under stationary conditions for an extended period. In meiosis, centromere clusters disintegrated before the emergence of the earliest precursors of the synaptonemal complex. Evidence for the contribution of centromere clustering to other aspects of suprachromosomal nuclear order, in particular the vegetative association of homologous chromosomes, is provided, and a possible supporting role in meiotic homology searching is discussed.
In dividing cells, centromeres congregate during early metaphase at the equator of the cell and cluster at prometaphase when they all become attached to the spindle apparatus which extends between the two opposite poles. At anaphase, centromeres disjoin and move to the poles with chromatids and telomeres dragging behind. Thus, at the telophase-G1 transition the centromeres and distal telomeres occupy opposite positions in the newly formed nucleus. The maintenance of this centromere– telomere polarization during the subsequent S and G2 stages of the interphase and even into prophase is a matter of debate. Rabl (1885) first reported on a polarized arrangement of interphase chromosomes in nuclei of salamander larvae, which thereafter became known as the Rabl-orientation. Subsequent investigations disclosed the Rabl-orientation in a variety of organisms. It was studied in most detail in Drosophila, where salivary gland nuclei were three-dimensionally reconstructed and the course of polytene chromosomes traced (Mathog et al., 1984). It was found that chromosomes on the whole show the Rabl-orientation, but follow a meandering path between the centromeric and telomeric pole with some chromosomes even looping back (Marshall et al., 1996 and references therein). Other examples from the animal kingdom that display aspects of a chromosome polarization are the embryonic cells of Ascaris (Boveri, 1909), fibroblasts of the tree shrew, Tupaia belangeri (Haaf and Ward, 1995a ), the lymphocytes of the Indian muntjac (Sperling and Lüdtke, 1981), and Chinese hamster fibroblasts (Cremer et al., 1982). However, most reports on Rabl-orientation stem from plants, where it is often visible as the polarized distribution of heterochromatin bands (for review see Avivi and Feldman, 1980; Fussell, 1987).
On the other hand, Houben et al. (1995) observed centromere clustering indicative of Rabl-orientation only in a few (presumably G1) interphase nuclei of field beans. Likewise, in human cells that are particularly well studied with respect to nuclear organization (see Cremer et al., 1993), evidence for Rabl-orientation is only sporadic. Manuelidis and Borden (1988, and references therein) found that centromeres rapidly disperse at the end of anaphase in human tissues. Moroi et al. (1981) and Earnshaw et al. (1987) did not find evidence for a Rabl-orientation in mammalian cell nuclei. Haaf and Schmid (1989) showed that although centromeres in human tumor cells were nonrandomly positioned close to the nuclear membrane during interphase, there was no clear evidence for a Rabl-like orientation of chromosomes. Moreover, the distinct domains occupied by individual chromosomes, visualized by chromosome painting in interphase with multiple color probes, do not show parallel alignment that would be indicative of Rabl-orientation (Dauwerse et al., 1992). In mouse lymphocytes, Vourc'h et al. (1993) found a non-Rabl chromosomal organization, which was also observed in mouse spermatogonia (Scherthan et al., 1996) and mouse Leydig cells (Scherthan, H., unpublished observations). The general picture that emerges from these conflicting reports is that in different organisms or in different tissues within an organism, anaphase polarization of chromosomes may be preserved to a different extent at interphase.
However, even in the absence of the Rabl-orientation, centromere clustering does occur. In human lymphocytes, Weimer et al. (1992) found that centromeres were located at the periphery of nuclei in the G0 stage and were clustered in ∼15 spots. When reinitiation of the mitotic cycle was stimulated with phytohemagglutinin, clusters were dissolved and centromeres moved towards the nuclear interior at S phase and adopted a near-random distribution at G2 (see also Ferguson and Ward, 1992). After mitosis, centromeres again showed a random distribution throughout the second interphase after stimulation. At no point was a prominent Rabl-orientation present. Likewise, clustering of centromeres into fewer spots than chromosomes present was described for human fibroblasts (Bartholdi, 1991), human and mouse sperm nuclei (Zalensky et al., 1995; Haaf and Ward, 1995b ), and mouse cells (Hsu et al., 1971; Rae and Franke, 1972; Brinkley et al., 1986). This seems to indicate that not only anaphase chromosome polarization but also active positioning at interphase govern chromosome arrangement within nuclei (see Marshall et al., 1997).
Since there are several reports on nonrandom distribution of chromosomes or chromosomal regions in yeast (Klein et al., 1992; Gilson et al., 1993; Guacci et al., 1994, 1997; Loidl et al., 1994; Weiner and Kleckner, 1994; Gotta et al., 1996), we wanted to investigate whether there exists a Rabl-like chromosomal arrangement in yeast nuclei and whether it could contribute to other observed aspects of suprachromosomal nuclear organization in this organism. We took advantage of the possibilities in yeast to obtain synchronized cultures enriched with specific stages of the mitotic cell cycle and to induce meiosis. By fluorescence in situ hybridization to spread cells in time course experiments, we followed the formation and resolution dynamics of specific chromosomal arrangements throughout the cell cycle.
Materials and Methods
Yeast Strains
Nuclei were obtained from the diploid Saccharomyces cerevisiae strain SK1 (Kane and Roth, 1974) at logarithmic growth and stationary phases, as well as meiotic stages. To study nuclei at specific stages of the cell cycle, cultures of both a haploid and a diploid strain were synchronized with α-factor, a yeast mating pheromone. Yeast cells that express the MAT a mating type allele only, are arrested at G1 when exposed to α-factor and resume growth upon removal. The haploid strain (4202-15-3a, kindly provided by L. Hartwell, University of Vienna, Vienna, Austria) was MAT a, ade2-1(ochre), his4-580(amber), lys2(ochre), trp1(amber), tyr1(ochre), SUP4-3(ts amber suppressor), bar1-1. The mutation in BAR1 prevents recovery from arrest after extended exposure to α-factor. The diploid strain (no. 183, kindly provided by F. Klein, Fred Hutchinson Cancer Research Center, Seattle, WA) was a derivative of SK1 with the genotype MAT a/MAT a, ho:: LYS2/ho::LYS2, lys2/lys2, leu2::hisG/leu2::hisG, his4/his4, ura3/ura3. In addition, a variety of common laboratory strains of different origins were used to verify the generality of the phenomenons reported here.
Cell Culture and Preparation
For logarithmic and stationary phase cells, cultures of various strains were grown in YPD. Cell densities were determined and samples containing 4 × 107 cells were collected at appropriate times. For synchronized cultures, haploid bar1 or diploid MAT a/MAT a cells were grown to a density of ∼107 cells/ml in 50 ml YPD, and α-factor (15 μg/ml; Sigma Chemical Co., St. Louis, MO) was added. After 3 h exposure to α-factor, a 4.5-ml aliquot of cells was removed and put on ice. From the remaining culture, α-factor was removed by pelleting and washing cells twice with an excess of YPD. Thereafter, cells were resuspended in YPD at 30°C. At subsequent 15-min intervals, 4.5-ml aliquots of the cell culture were removed and stored on ice. (An alternative approach to discriminate between G1 and G2 nuclei on the basis of duplication of fluorescence signals on chromosome IV due to their replication status (see Selig et al., 1992) was abandoned because of the ambiguous appearance of signals.)
For meiotic cells, cultures were grown in presporulation medium to a density of 2 × 107 cells/ml and then transferred to sporulation medium (2% KAc) at a density of 4 × 107 cells/ml (Roth and Halvorson, 1969). Aliquots from the sporulating cultures were taken at regular intervals and immediately put on ice.
Cells were spheroplasted with Zymolyase 100T (140 μg/ml; Kirin Brewery Co. Ltd., Tokyo, Japan) in 0.8 M sorbitol supplemented with 10 mM DTT. Spheroplasting was terminated by adding 10 vol of ice-cold 1 M sorbitol. Cells were pelleted and resuspended at a concentration of 4 × 108 cells/ml. This suspension was then mixed with detergent and fixative on a slide for spreading the cells according to Loidl et al. (1991). For detailed protocols see also Loidl et al. (1998).
For the preparation of unspread nuclei, we embedded cells in a polyacrylamide layer on the slide according to Bass et al. (1997). Cells were spheroplasted and a cell suspension was prepared as above. A 5% acrylamide solution was prepared by diluting a 30% activated acrylamide stock with 4% paraformaldehyde/3.4% sucrose solution. 5 μl of the cell suspension was added to 80 μl of the acrylamide/fixative solution. The mixture was dropped onto a slide, covered with a 24 × 60-mm coverslip and sealed with silicon gel. After polymerization/fixation for 30 min at room temperature the coverslip was removed, and after rinsing with 4× SSC + 0.1% Tween 20 for at least 5 min the preparation was subject to the hybridization procedure as below. Care was taken to avoid drying between the various incubation steps.
DNA Probes and Labeling
11 λ-clones containing sequences of 11 of the 16 yeast centromeres were obtained from the American Type Culture Collection (no. 70028, 70300, 70385, 70464, 70549, 70583, 70597, 70610, 70622, 70641, 70667; ATCC, Gaithersburg, MD). λ-DNA was prepared according to a minilysate protocol (Davis et al. 1986) and further amplified using the degenerate oligonucleotide-primed PCR protocol by Telenius et al. (1992). The PCR products were used as centromere-specific probes for fluorescence situ hybridization (FISH).1 The remaining five centromeres of chromosomes I, III, VIII, XII, and XIII were probed with cosmid clones of centromeric or centromere-near regions (70893, 70889, 71205, 71055, 70921; ATCC).
Plasmid pEL42H10-4.8HR containing a fragment with the conserved core of the subtelomeric Y′ element, and clone pEL113H containing a 2-kbp fragment harboring the conserved core of the subtelomeric X element (Louis et al., 1994; kindly provided by E.J. Louis, John Radcliffe Hospital, Oxford, UK) were used as probes to yeast telomeres (see also Gotta et al., 1996). To enhance signal intensity and to create a pantelomeric DNA-probe, we mixed the X and Y′ probes in equimolar amounts before labeling. Specificity of probes was confirmed by FISH to condensed pachytene chromosomes where (80% of telomeres showed readily detectable signals (not shown). To label the end of the long arm of chromosome IV (IVR) and the short arm of chromosome III specifically, probes 71013 (ATCC; cosmid) and 70303 (λ) were used.
The composite pancentromeric DNA probe (a mixture of PCR products and cosmids) was labeled with Cy3-conjugated dUTP (Amersham Corp., Arlington Heights, IL) and the pantelomeric probe was labeled with biotin-16-dUTP (Boehringer Mannheim Corp., Indianapolis, IN) using a nick translation kit according to the instructions of the supplier (GIBCO BRL, Gaithersburg, MD). Specific regions on chromosomes III and IV were labeled with biotin or both, Cy3 and biotin, depending on the experiment.
FISH
FISH was carried out as described previously (Scherthan et al., 1992) with slight modifications. In brief, DNA probes labeled with biotin or Cy3 were dissolved at 30 ng/μl in hybridization mixture (50% formamide, 2× SSC, 10% dextran sulfate, 1 μg/μl salmon sperm carrier DNA) and put onto the slide under a coverslip. Preparations were denatured for 5 min at 74°C and hybridized for 48 h at 37°C (Steinmüller et al., 1993). Subsequently, biotinylated probes were detected using FITC-conjugated avidin (Sigma Chemical Co., St. Louis, MO). Finally, preparations were embedded in antifade medium (Vector Laboratories Inc., Burlingame, CA) containing 0.5 μg/ml DAPI (4′6-diamidino-2-phenylindole) as DNA-specific counterstain.
Microscopic Evaluation
Preparations were evaluated using Zeiss Axioskop epifluorescence microscopes equipped with a double-band-pass filter for simultaneous excitation of red and green fluorescence and single band pass filters for excitation of red, green, and blue (Chroma Technologies, Brattleboro, VT). Images of high magnification and resolution were obtained using cooled black and white CCD cameras controlled by the ISIS fluorescence image analysis system (MetaSystems, Altlussheim, Germany) and IPLab Spectrum software (Scanalytics, Fairfax, VA), respectively. Hundred well-hybridized nuclei were scored for each time point. Nuclei were preselected on the basis of an undisrupted, homogeneous appearance in the DAPI-stained image. Grossly jagged or deformed nuclei were excluded from analysis.
Results
Centromere Clustering Is Frequent in Logarithmic, but Rare in Stationary Culture Cells
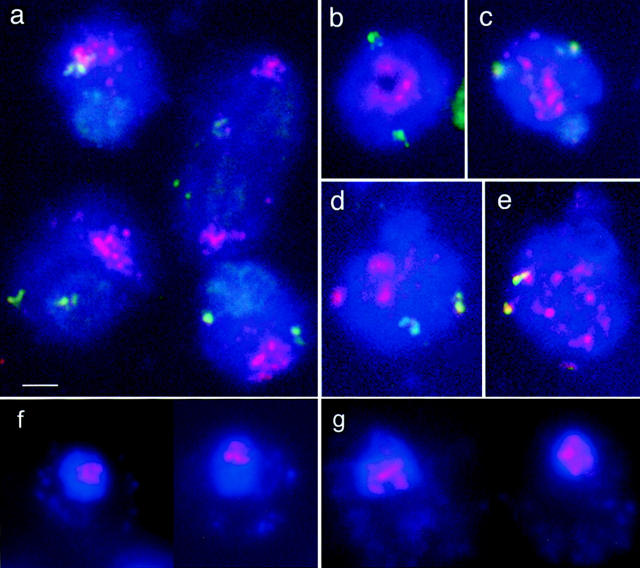
The nuclear distribution patterns of centromeres were analyzed under different growth conditions and at different stages of the mitotic cell cycle as well as in meiotic prophase in the budding yeast, S. cerevisiae. In spread nuclei we found a high incidence of centromere clustering throughout interphase in a variety of strains (Figs. 1 and 2). Nuclei where the distribution of centromeric FISH signals was restricted to <50% of the total area of the DAPI-stained spread nucleus and nuclei with no more than four separate patches of fused centromere signals were classified as showing centromere clustering. In roughly half of these nuclei, centromeric FISH signals occupied even ⩽25% of the DAPI stained nuclear chromatin. In some cultures of strain SK1, ⩽95% of nuclei showed clustered signals with a pancentromeric probe. Sometimes, centromeres were arranged in a ring (Fig. 1 b) whose significance we have not yet fully explored. A subset of nuclei showed a typical anaphase polarization of centromeres and telomeres (Figs. 1 a and 2, e–g).
Figure 1.
Representative nuclei of SK1 cells from (a, b, f) logarithmic growth and (c–e, g) stationary phase culture. (a–e) Spread nuclei; (f and g) intact nuclei. (Red) Centromeres. Telomeric region of chromosome IVR, green. (Blue) DAPI-stained chromatin. (a) Nuclei show centromere clusters near the periphery. Telomeres are dispersed within the nuclear chromatin. A slight preference for accumulation of telomere signals in the centromere-distant domain of the nucleus was noted (see text). The nucleus right on top is at anaphase with centromeres at the two opposite poles of the elongated nucleus and telomeres IVR in between. (b) Plan view of the centromeric pole of a nucleus showing centromeres arranged in a ring. (c–e) Nuclei displaying aspects of the progressive reduction of centromere clustering in a stationary culture. (f and g) Examples of intact nuclei from three-dimensionally embedded spheroplasts. Cells from a logarithmic culture show centromere clusters (f) and cells from a stationary culture show dispersed centromeres (g). Notice the ring-like arrangement of centromeres in the left nucleus in f which is similar to b. Nuclei are delineated in strong blue, spheroplasts are lightly blue due to DAPI staining of the mitochondrial DNA. Bar, 2 μm.
Figure 2.
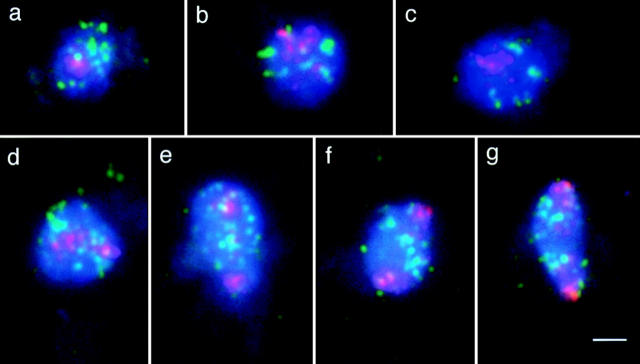
FISH of centromeric (red) and telomeric (green) regions in spread mitotic nuclei of the haploid bar1 strain. (Blue) DAPI-stained chromatin. (a–c) Interphase nuclei with clustered centromeric regions. Telomeres are apparently randomly distributed in the nucleus in a, whereas in b and c there is some association of telomere signals (see text). (d–g) Mitotic nuclei showing different degrees of separation of centromere clusters (pictures taken at t = 75 min of a mitotic time course), probably representing metaphase and anaphase stages. (d) The centromere cluster is split into two equal-sized patches. (e and f) The clusters consisting of the centromeres of chromatids are separated further and most telomeres are assembled between them. (g) The nucleus is oblong with the centromeres at the most distant poles, and the telomeres are separated to the two halves of the nucleus. Note that the constriction in the middle of the dividing nucleus, that is typical of anaphase/telophase in intact cells (for example see Byers, 1981), is not retained after spheroplasting. Bar, 2 μm.
Since in spreads some spatial information is lost and artefacts may be produced by drying and spreading forces, we performed FISH on the intact nuclei of spheroplasts embedded in polyacrylamide. Like in spread cells, we observed tight centromere clustering in a high proportion of intact nuclei but their small size precluded a detailed analysis (Fig. 1, f and g). For the benefit of enhanced resolution the experiments reported below were performed with spread nuclei.
Frequencies of centromere clustering were compared between logarithmically growing and stationary cultures. Whereas in a SK1 culture at 1 × 107 cells/ml, 95% of the cells displayed centromere clusters (Fig. 1, a and b); clustering was reduced to 37% after the culture had been left for 48 h at 2.6 × 108 cells/ml (Fig. 1, c–e). Likewise, in strain no. 183, clustering was 93% in a culture with 5 × 106 cells/ml and was reduced to 36% at 1.1 × 108 cells/ml after 24 h. (In both experiments 100 nuclei were checked for each time point.) We conclude that centromere clusters become unstable in the increasing intervals between mitoses in dense, slowly growing cultures although clustering did not completely disappear even after several days (not shown).
Centromere–Telomere Polarization
To study the relative distribution patterns of centromeres and telomeres we performed two sets of experiments. In one we labeled centromeres and telomeric regions of all chromosomes. In the other we labeled all centromeres and the telomeres of the long arm of chromosome IV (IVR) specifically.
In spread nuclei where centromeres and all telomeres were simultaneously labeled with Cy3 (red) and FITC (green), we found that centromeres usually were organized in one big cluster whereas the number of telomeric FISH signals (in the haploid strain) varied between ∼10 and >20 (Fig. 2, a–c). From immunolabeling and FISH in structurally preserved yeast cells it is known that telomeres are clustered in a limited number of foci, preferentially positioned near the nuclear envelope (Klein et al., 1992; Gotta et al., 1996). The detection in our preparations of fewer telomeric signals than telomeres present (32 in the haploid) may reflect this telomere clustering, but since telomere–telomere associations are partially lost in detergent-spread nuclei (Klein et al., 1992; Palladino et al. 1993), it was only weakly expressed.
In the experiment with the pancentromeric and chromosome IVR telomere-specific probes we found that centromeres and telomeres tend to occupy opposite regions of the nucleus. In 100 out of 110 arbitrarily selected spread nuclei of a logarithmic growth phase culture of the SK1 diploid strain, centromere clusters were located at the periphery. This suggests that in vivo centromeres are assembled near the nuclear membrane also. Of the 200 chromosome IVR telomere signals in these nuclei, 82 (41%) were in the centromere-distal third of the nucleus, 64 (32%) in the median third and 54 (27%) were in the proximal third. If chromosome arm IVR (which is long enough to span the diameter of the nucleus, see Discussion) strictly followed the Rabl-orientation, its telomere should be located in the centromere-distal domain of the nucleus. The preferential, but not exclusive, position of telomeres in the centromere-distal domain suggests that chromosome arms by and large follow a Rabl-like orientation but may sometimes deviate from the ideal path (see also Marshall et al., 1996).
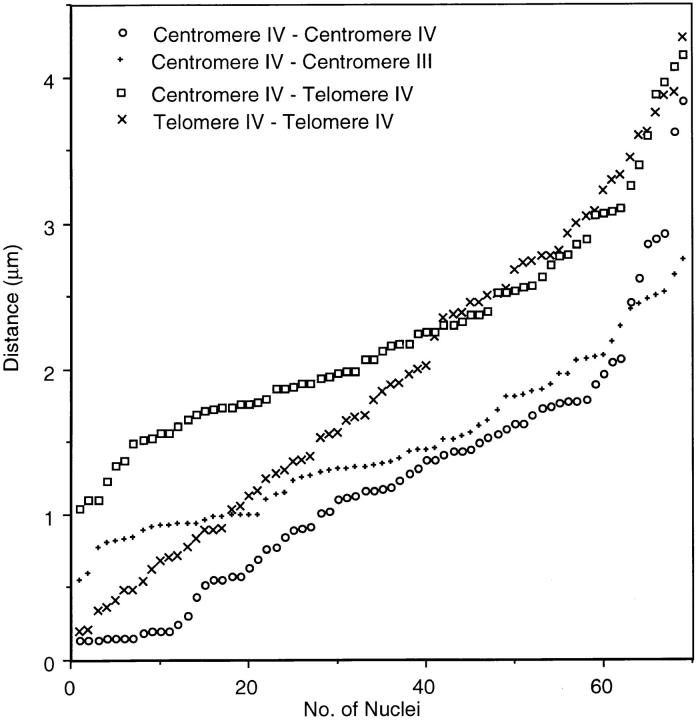
In 69 diploid nuclei we measured distances between homologous and nonhomologous centromeric regions (on chromosomes III and IV) that are both included in the centromere cluster, and between these centromeric regions and the telomeric regions on IVR (Fig. 3). The mean distances between centromeres and telomeres IVR were notably larger than the distances between centromeres (Fig. 3), which confirms that telomeres of IVR tend to occupy areas distant from the centromere cluster. Interestingly, distances between the centromeres of homologous chromosomes IV were slightly smaller than between the centromeres of III and IV. Whether this is due to specific homologous interactions (see Discussion) will be further investigated. Homologous telomeres IVR were on the average closer together than centromeres and telomeres IVR, which might reflect telomere association (see above). However, this feature appears to be lost in strongly spread nuclei.
Figure 3.
Pairwise distances between homologous chromosome regions within the centromere cluster (Centromeres IV), nonhomologous chromosome regions within the centromere cluster (Centromeres IV – Centromere-near III), the centromere and the telomere of the long arm of chromosome IV, and between the homologous telomeres IV in the diploid strain SK1. The distances between the centers of FISH signals were measured in 69 nuclei which were differentially stained for these three regions. For nonhomologous signals all four possible distances were pooled for each nucleus. All measured distances were independently plotted in increasing order, i.e., symbols on the same ordinate do not necessarily represent distances in one and the same nucleus. This format of presentation adopted from Weiner and Kleckner (1994) is highly suitable for accentuating minute differences in average distances.
Centromeres Are Clustered Throughout the Mitotic Cell Cycle
To study the redistribution of centromeres and telomeres during mitosis and substages of interphase, mitotic time course experiments were conducted. To this end, cultures of both a diploid and a haploid strain were synchronized with α-factor (see Materials and Methods). In the diploid MAT a/MAT a strain (no. 183) 94% of cells (n = 200) were without a bud after 3 h exposure, which indicates high synchronization at G1 (Sprague, 1991). Samples taken at 15-min intervals from t = 0 to t = 105 minutes after release from the arrest, showed between 81 and 90% centromere clustering (n = 100) with no tendency towards the higher or lower frequency over time (Table I).
Table I.
Frequency of Nuclei with Clustered Centromeres (in Percent) Over a Mitotic Time Course in the Diploid MATa/ MATa and the Haploid bar1 Strain Synchronized with α-Factor
| Minutes after release from α-factor | 0 | 15 | 30 | 45 | 60 | 75 | 90 | 105 | 120 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diploid | Percent centromere clusters | 83 | 85 | 89 | 90 | 81 | 90 | 82 | 88 | ND | ||||||||||
| Haploid | Percent centromere clusters | 81 | 77 | 80 | 79 | 83 | 68 | 78 | 81 | 80 | ||||||||||
| Percent anaphases | 0 | 0 | 0 | 0 | 0 | 16 | 10 | 1 | 0 | |||||||||||
100 nuclei were evaluated for each time point in both the diploid and the haploid strain. In the haploid, the presence of telomere signals allowed the identification of anaphases. Nuclei were classified as being at anaphase when two centromere clusters were present at opposite peripheral positions with most telomere signals between them (compare Fig. 2, f with g). The frequency of centromere clustering was roughly constant for all time points except for t = 75 min in the haploid where a large proportion of nuclei showed anaphase configurations instead.
A time course in the haploid bar1 strain showed a slightly lower incidence of centromere clustering of ∼80% (Table I) that could relate to the different strain background or experimental conditions. In this experiment dividing nuclei (Fig. 2, d–g) appeared primarily at time points t = 75 min and t = 90 min (Table I). Mitoses were characterized by the splitting of centromere clusters into two roughly equal halves that occupied opposite poles. In both strains, the frequencies of centromere clusters were roughly the same at timepoints before and after the occurrence of mitoses, which indicates that there is no obvious difference between G1 and G2 nuclei with respect to centromere clustering.
Centromere Clusters Are Rapidly Dissolved at Meiotic Prophase
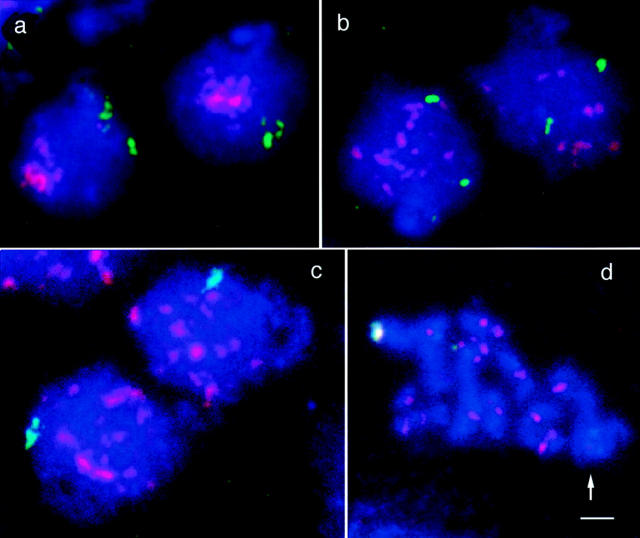
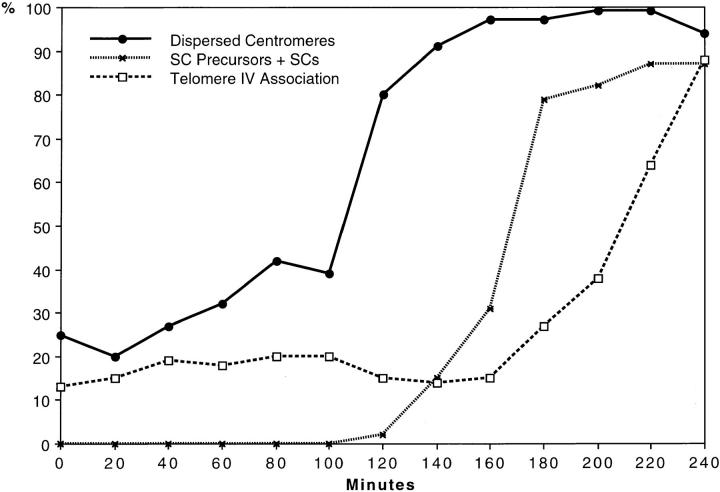
A meiotic time course with samples taken at 20-min intervals showed that centromere clustering started to decrease soon after the transfer to sporulation medium. After 120 min, the frequency of nuclei with clustered centromeres had dropped from 61 to 20% (Fig. 4). The resolution of clusters occurred in advance of the time when precursors of the synaptonemal complex (SC) first emerged (Fig. 5). After 160 min in sporulation medium 97% of nuclei were without centromere clusters, but the frequency of nuclei with SCs or SC precursors was only 31%. The fusion of FISH signals from chromosome IVR telomeric regions, which indicates homologous pairing, started to increase over mitotic levels only after 180 min in sporulation medium (Fig. 5). The earlier onset and completion of the resolution of clusters as compared with the formation of SC precursors and homologous pairing suggests that resolution of centromere clusters reflects a structural reorganization of the nucleus in preparation for synapsis. It is notable that during the first 100 min in meiosis there was a relatively high frequency (∼20%) of associated FISH signals of telomere IVR. Around t = 140 min in meiosis it was slightly lower (14%). This decrease in association between homologous regions coincides with the dramatic increase of nuclei that have lost centromere clustering between t = 100 and t = 160 min (Fig. 5). Only at later timepoints, the fusion of telomere IVR signals rises again as a consequence of synapsis. It may be concluded that the restructuring of nuclear architecture that leads to the loss of centromere clustering disrupts also the spatial relationship that exists between homologous regions in vegetative nuclei (see Discussion). This reduction in homologous associations matches well with the transient decrease in pairing at around t = 120 min in sporulation medium, which was observed by Weiner and Kleckner (1994) and was interpreted as the disruption of presynaptic pairing interactions by chromosome replication.
Figure 4.
Change in the nuclear architecture upon transfer to sporulation medium. (a) Early in meiosis, centromere (red) clustering is striking and telomeric regions on the long arms of homologous chromosomes IV (green) tend to be located near the nuclear periphery opposite to the centromere cluster. As can be seen from the two examples depicted in a, this polarization may bring about a closer than random association between homologous telomeres, since their distribution is restricted to a relatively small domain of the nucleus. At later time points nuclei with scattered centromeres become predominant. In early nuclei of this type, homologous telomeres IV produce separate signals (b), whereas later the telomeric signals fuse (c) in the course of meiotic pairing. For the frequencies of the various structural aspects at different time points in the meiotic culture see Fig. 5. (d) At pachytene bivalents are condensed. The red FISH signals indicate centromeres of individual bivalents. The green spot marks the end of the synapsed long arms of chromosome IV. The region to the right, which is devoid of centromere signals, is the nucleolus (arrow). Chromatin is stained blue with DAPI. Bar, 2 μm.
Figure 5.
Meiotic time course showing the increase of frequency of nuclei with dispersed centromeres (i.e., loss of centromere clustering) and the appearance of SCs and their precursors (short axial elements and short synapsed segments). For each time point (samples taken at 0 to 240 min in sporulation medium in 20 min intervals) 100 nuclei were analyzed by FISH and light microscopy of silver-stained preparations (for example see Loidl et al., 1991) for centromere clustering, homologous associations of telomeres IVR, and presence of SCs or SC-precursors. The increase in the frequency of nuclei without clustered centromeres precedes the first appearance of SC-precursors at ∼120 min. Still later (180 min) the association of homologous chromosomal regions (telomere-near sites on chromosome IVR) above mitotic background levels indicates the onset of synapsis.
Discussion
Does Centromere Clustering at Interphase Reflect Anaphase Chromosome Orientation or Another Kind of Nuclear Organization?
We have studied centromere distribution in both intact and gently spread nuclei. Spreading was performed to improve the spatial resolution of nuclear structures. FISH with a pancentromeric probe showed that centromere clustering is a prominent feature of interphase nuclear architecture in rapidly dividing budding yeast cells. Centromere clusters generally occupied a small domain at the periphery of the nucleus, from which the chromosome arms projected.
Recently, Guacci et al. (1997) had also inferred a cell cycle–dependent clustering of centromeres from the closer than random spatial association of the centromeres of three chromosomes. Whereas it is reasonable to assume that centromere clustering is a consequence of anaphase chromosome polarization, and thus an aspect of the Rabl-orientation, it would probably be randomized by Brownian motion if it was not fixed by the anchoring of chromatin to the nuclear matrix or the nuclear envelope (Marshall et al., 1997). In addition or alternatively to residual anaphase orientation, centromere clustering might be generated by the active positioning of chromatin by a motor-like activity within the interphase nucleus (see Marshall et al., 1997).
Centromere clustering was also reported in the fission yeast Schizosaccharomyces pombe where >80% of nuclei at G2 phase showed clusters (Funabiki et al., 1993). In this organism centromeres cluster adjacent to the spindle pole body, which is evidence for the origin of centromere clustering in the course of the anaphase movement. The spatial relationship between centromeres and spindle pole body is preserved during a rotation of nuclei at cytokinesis when spindle pole bodies move out of the axis of the former division spindle. This concerted movement of centromeres and spindle pole body may indicate a physical connection of the centromeres with the cytoskeleton, which is maintained beyond mitosis. Also for S. cerevisiae it may be assumed that centromeres are kept in place by their permanent attachment to the spindle pole body, since it has been shown that nuclear microtubules are present throughout the entire cell cycle (Byers and Goetsch, 1975), and there is evidence that centromeres and spindle pole bodies colocalize during interphase (Guacci et al., 1997). The identification of a mutation (crm1) that affects centromere clustering in fission yeast (Adachi and Yanagida, 1989; Funabiki et al., 1993) provides additional evidence for the existence of a structure or activity that helps to maintain centromere clustering after its origin during anaphase polarization, or creates centromere clustering de novo during interphase.
Our experiments showed that in G0 nuclei of stationary cultures, when cells have not divided for several hours, the clustering of centromeres is reduced. This could either mean that anaphase centromere orientation simply disintegrates in the absence of a stabilizing structure or mechanism (e.g., stable attachment to the nuclear envelope and/or the nuclear matrix), or that any activity that might create centromere clusters during interphase does not persist in stationary phase cells. Likewise, it was observed in Drosophila that during the short intervening interphases of mitotically highly active embryonic cells, chromosomes remain in an extended configuration. However, in later stages of development when interphase is much longer, physical interactions between centromere-near and -distal chromosomal sites do occur (Dernburg et al., 1996). These authors ascribed this effect to the relaxation of chromosomes from the Rabl-conformation during long interphases.
Apart from being a mere reflection of spatial constraints within the nucleus, centromere clustering may be functionally important at metaphase. In yeast mitosis centromeres do not assemble at the equator of the dividing cell (the so-called metaphase plate; Straight et al., 1997). The clustering of centromeres, however, guarantees that centromeres are not scattered all around the nucleus by the time the spindle is formed. It is possible that both the metaphase plate in higher eukaryotes and the centromere cluster in yeast (although not necessarily at an equidistant position to the two spindle poles, see Straight et al., 1997) serve to facilitate the attachment of chromosomes to the spindle.
Centromere Clustering and Vegetative Pairing
Evidence regarding the existence of vegetative or somatic homologous pairing in yeast and higher eukaryotes other than dipterans is controversial (for review and contrasting views see Stack and Brown, 1969; Avivi and Feldman, 1980; Comings, 1980; Lacadena et al., 1983; Therman and Denniston, 1984; Hadlaczky et al., 1986; Hilliker and Appels, 1989; Loidl, 1990; Haaf and Schmid, 1991; Kleckner and Weiner, 1993; Haber et al., 1996; Henikoff, 1997). Vegetative pairing could reflect the fusion of similar arrays of transcription units on homologous chromosomes due to the demand to share transcription factors and polymerases (Cook, 1997). It has also been speculated that vegetative pairing could have evolved from the need of G1/G0 cells to perform recombinational repair in the absence of sister chromatids and that meiotic pairing would be facilitated by the existence of this type of pairing or association in vegetative or premeiotic cells (see Kleckner and Weiner, 1993). In fact, in yeast cultures which had been grown in presporulation medium there is a remarkably high incidence of homologously associated FISH signals at t = 0 of meiosis (Loidl et al., 1994; Weiner and Kleckner, 1994). In this case, however, one could argue that cells are stimulated by the suboptimal growth conditions in the poor presporulation medium to initiate homology searching as a prelude to meiosis. Keeney and Kleckner (1996), however, reported that also in vegetatively growing nuclei homologous chromosomes appeared to be paired. Based on the observation that DNase I sensitivity at a nuclease-hypersensitive site was higher when the same allele was present on the homologous chromosome, they suggested that local homologous interactions would bring about physical contact. Similarly, it was shown by LaSalle and Lalande (1996) that oppositely imprinted sites on homologous chromosomes associate in human lymphocytes. Also we found a slight tendency for homologous regions to be more closely associated than expected on the basis of random distribution in vegetative nuclei (see Fig. 3; Jin, Q.-w., and J. Loidl, unpublished observations). This might be due to specific homologous interactions, but it is also possible that a Rabl-like orientation causes or at least contributes to homologous associations by assigning specific nuclear positions to chromosome arms dependent on their size.
In yeast, interphase chromatin was estimated to be compacted by a factor of ∼80 compared with naked DNA (Guacci et al., 1994). Taking an estimate of 2.9 kb/μm DNA, one can calculate the interphase lengths of all chromosome arms (based on chromosome sizes given by Cherry et al., 1997). Even if these parameters are somewhat uncertain, it seems that only the ten longest of the 32 yeast interphase chromosome arms would have to be folded further to be accommodated in the nucleus whose diameter is ∼2 μm. The medium-sized and short arms of the complement, on the other hand, would not extend from the centromeric to the opposite pole of the nucleus but only to median regions. These arms would be positioned at the nuclear periphery as telomeres tend to associate with the nuclear envelope (Klein et al., 1992; Gotta et al., 1996), and would consequently displace longer arms toward the interior of the nucleus. This nuclear order could impose a topological constraint by which chromosomes of same lengths, i.e., homologues, would tend to occupy similar positions in the interphase nucleus and be closer together than if their distribution was random. A similar tendency of chromosomes to adopt non-random positions merely by following physical constraints is also illustrated by their size-dependent assortment in human metaphase plates (Mosgöller et al., 1991). Thus, self-organization rather than specific control may be a considerable factor in establishing even seemingly complex patterns of order.
Nuclear Architecture and Meiotic Chromosome Pairing
It is a much-debated question whether meiotic chromosome pairing benefits from some kind of premeiotic chromosomal disposition. Some authors suggested that vegetative pairing or other modes of non random chromosomal arrangement reduce the expenditure on meiotic homology search (see Stack and Brown, 1969; Loidl, 1990; Kleckner and Weiner, 1993). Fussell (1987) proposed that the Rabl-orientation carries over into meiosis, which would minimize the movement of chromosomes to form a bouquet (i.e., an arrangement by which all telomeres assemble in a small region near the nuclear envelope) and thus to initiate synapsis. In maize, Bass et al. (1997) found a centromere– telomere grouping that is consistent with a Rabl-configuration in premeiotic mitosis. However, this grouping is lost as cells pass through premeiotic interphase and leptotene. Thus, the bouquet forms de novo during meiotic prophase. In the mouse, Scherthan et al. (1996) did not observe centromere–telomere polarization in several hundreds of spermatogonia and also reached the conclusion that the Rabl-configuration does not contribute to bouquet formation and homology search. It has been reported that a bouquet stage exists in the meiosis of budding yeast (Dresser and Giroux, 1988; Trelles-Sticken, E., and H. Scherthan, unpublished observations), but frequency and precise timing of its occurrence have not yet been determined and are currently under investigation by us. It is well possible that the observed loss of centromere clustering in meiotic nuclei (Fig. 5) is due to bouquet formation. When telomeres converge in a small area near the nuclear surface, the centromere cluster will be disrupted as arm lengths will dictate the positioning of centromeres relative to this region. A similar transition at which centromere clusters dissolve whilst telomeres aggregate, was recently described for early meiotic prophase of wheat (Aragón-Alcaide et al., 1997).
Studying the relative timing of the resolution of centromere clusters and the appearance of the bouquet will allow us to determine whether the Rabl-like orientation in yeast is directly transformed into the bouquet, or whether a more complex rearrangement of centromeric versus telomeric attachments to the nuclear envelope (Scherthan et al., 1994, 1996; Bass et al., 1997; Chikashige et al., 1997) precedes meiotic chromosome pairing.
Acknowledgments
We are grateful to Franz Klein for valuable comments on the manuscript.
This work was supported by grant no. 58202 from the Austrian Fund for the Advancement of Scientific Research (FWF). H. Scherthan received support from the Deutsche Forschungsgemeinschaft (Sche 350/8-1).
Abbreviations used in this paper
- DAPI
4′6-diamidino-2-phenylindole
- FISH
fluorescence in situ hybridization
- SC
synaptonemal complex
Footnotes
Please address all correspondence to J. Loidl, Institute of Botany, University of Vienna, Rennweg 14, A-1030 Vienna, Austria. Tel.: 43 1 79794 170. Fax: 43 1 79794 131. E-mail: josef.loidl@univie.ac.at
References
- Adachi Y, Yanagida M. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene crm1 +which encodes a 115-kD protein preferentially localized in the nucleus and its periphery. J Cell Biol. 1989;108:1195–1207. doi: 10.1083/jcb.108.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón-Alcaide L, Reader S, Beven A, Shaw P, Miller T, Moore G. Association of homologous chromosomes during floral development. Curr Biol. 1997;7:905–908. doi: 10.1016/s0960-9822(06)00383-6. [DOI] [PubMed] [Google Scholar]
- Avivi L, Feldman M. Arrangement of chromosomes in the interphase nucleus of plants. Hum Genet. 1980;55:281–295. doi: 10.1007/BF00290206. [DOI] [PubMed] [Google Scholar]
- Bartholdi MF. Nuclear distribution of centromeres during the cell cycle of human diploid fibroblasts. J Cell Sci. 1991;99:255–263. doi: 10.1242/jcs.99.2.255. [DOI] [PubMed] [Google Scholar]
- Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. JCell Biol. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri Th. Die Blastomerenkerne von Ascaris megalocephala und die Theorie der Chromosomenindividualität. Arch Zellforsch. 1909;3:181–268. [Google Scholar]
- Brinkley BR, Brenner SL, Hall J M, Tousson A, Balczon RD, Valdivia MM. Arrangements of kinetochores in mouse cells during meiosis and spermiogenesis. Chromosoma. 1986;94:309–317. doi: 10.1007/BF00290861. [DOI] [PubMed] [Google Scholar]
- Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces. Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 59–96.
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. . J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J.M., C. Adler, C. Ball, S. Dwight, S. Chervitz, Y. Jia, G. Juvik, T. Roe, S. Weng, and D. Botstein. 1997. Saccharomyces Genome Database. http://genome-www.stanford.edu /saccharomyces/. [DOI] [PMC free article] [PubMed]
- Chikashige Y, Ding D-Q, Imai Y, Yamamoto M, Haraguchi T, Hiraoka Y. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. . EMBO (Eur Mol Biol Organ) J. 1997;16:193–202. doi: 10.1093/emboj/16.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comings DE. Arrangement of chromatin in the nucleus. Hum Genet. 1980;53:131–143. doi: 10.1007/BF00273484. [DOI] [PubMed] [Google Scholar]
- Cook PR. The transcriptional basis of chromosome pairing. J Cell Sci. 1997;110:1033–1040. doi: 10.1242/jcs.110.9.1033. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C, Baumann H, Luedtke E-K, Sperling K, Teuber V, Zorn C. Rabl's model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments. Hum Genet. 1982;60:46–56. doi: 10.1007/BF00281263. [DOI] [PubMed] [Google Scholar]
- Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schröck E, Speicher MR, Mathieu U, Jauch A, Emmerich P, Scherthan H, Ried T, Cremer C, Lichter P. The role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harbor Symp Quant Biol. 1993;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- Dauwerse JG, Wiegant J, Raap AK, Breuning MH, van Ommen GJB. Multiple colors by fluorescence in situhybridization using ratio-labeled DNA probes create a molecular karyotype. Hum Mol Genet. 1992;1:593–598. doi: 10.1093/hmg/1.8.593. [DOI] [PubMed] [Google Scholar]
- Davis, L.G., M.D. Dibner, and J.F. Battey. 1986. Basic Methods in Molecular Biology. Elsevier, New York. 211–218.
- Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- Dresser ME, Giroux CN. Meiotic chromosome behavior in spread preparations of yeast. J Cell Biol. 1988;106:567–573. doi: 10.1083/jcb.106.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Sullivan KF, Machlin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield NF, Cleveland DW. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M, Ward DC. Cell cycle dependent chromosomal movement in pre-mitotic human T-lymphocyte nuclei. Chromosoma. 1992;101:557–565. doi: 10.1007/BF00660315. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussell, C.P. 1987. The Rabl orientation: a prelude to synapsis. In Meiosis. P.B. Moens, editor. Academic Press, Orlando. 275–299.
- Gilson ET, Laroche T, Gasser SM. Telomeres and the functional architecture of the nucleus. Trends Cell Biol. 1993;3:128–134. doi: 10.1016/0962-8924(93)90175-z. [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. . J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Schmid M. Centromeric association and non-random distribution of centromeres in human tumour cells. Hum Genet. 1989;81:137–143. doi: 10.1007/BF00293889. [DOI] [PubMed] [Google Scholar]
- Haaf T, Schmid M. Chromosome topology in mammalian interphase nuclei. Exp Cell Res. 1991;192:325–332. doi: 10.1016/0014-4827(91)90048-y. [DOI] [PubMed] [Google Scholar]
- Haaf T, Ward DC. Rabl orientation of CENP-B box sequences in Tupaia belangerifibroblasts. Cytogenet Cell Genet. 1995a;70:258–262. doi: 10.1159/000134047. [DOI] [PubMed] [Google Scholar]
- Haaf T, Ward DC. Higher order nuclear structure in mammalian sperm revealed by in situ hybridization and extended chromatin fibers. Exp Cell Res. 1995b;219:604–611. doi: 10.1006/excr.1995.1270. [DOI] [PubMed] [Google Scholar]
- Haber JE, Leung WY. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci USA. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadlaczky GY, Went M, Ringertz NR. Direct evidence for the non-random localization of mammalian chromosomes in the interphase nucleus. Exp Cell Res. 1986;167:1–15. doi: 10.1016/0014-4827(86)90199-0. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Nuclear organization and gene expression: homologous pairing and long-range interactions. Curr Opin Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- Hilliker AJ, Appels R. The arrangement of interphase chromosomes: structural and functional aspects. Exp Cell Res. 1989;185:297–318. doi: 10.1016/0014-4827(89)90301-7. [DOI] [PubMed] [Google Scholar]
- Houben A, Guttenbach M, Kreβ W, Pich U, Schubert I, Schmid M. Immunostaining and interphase arrangement of field bean kinetochores. Chromosome Res. 1995;3:27–31. doi: 10.1007/BF00711158. [DOI] [PubMed] [Google Scholar]
- Hsu TC, Cooper JE, Mace ML, Jr, Brinkley BR. Arrangement of centromeres in mouse cells. Chromosoma. 1971;34:73–87. doi: 10.1007/BF00285517. [DOI] [PubMed] [Google Scholar]
- Kane SM, Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Kleckner N. Communication between homologous chromosomes: genetic alterations at a nuclease-hypersensitive site can alter mitotic chromatin structure at that site both in cis and in trans. . Genes Cells. 1996;1:475–489. doi: 10.1046/j.1365-2443.1996.d01-257.x. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Weiner BM. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harbor Symp Quant Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF-X, Schweizer D, Gasser SM. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM, Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- Lacadena, J.R., B. Jodar, and E. Ferrer. 1983. Suprachromosomal organisation: cytogenetical rationale and experimental evidence. In Kew Chromosome Conference II. P.E. Brandham, and M.D. Bennett, editors. George Allen & Unwin, London. 81–90.
- Loidl J. The initiation of meiotic chromosome pairing: the cytological view. Genome. 1990;33:759–778. doi: 10.1139/g90-115. [DOI] [PubMed] [Google Scholar]
- Loidl J, Nairz K, Klein F. Meiotic chromosome synapsis in a haploid yeast. Chromosoma. 1991;100:221–228. doi: 10.1007/BF00344155. [DOI] [PubMed] [Google Scholar]
- Loidl J, Klein F, Scherthan H. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 1994;125:1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl, J, F. Klein, and J. Engebrecht. 1998. Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. In Methods in Cell Biology. Vol. 53 Nuclear Structure and Function. M. Berrios, editor. Academic Press, San Diego. 257–285. [DOI] [PubMed]
- Louis EJ, Naumova ES, Lee A, Naumov G, Haber J. The chromosome end in yeast: its mosaic nature and influence on recombination dynamics. Genetics. 1994;136:789–802. doi: 10.1093/genetics/136.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L, Borden J. Reproducible compartmentalization of individual chromosome domains in human CNS cells revealed by in situhybridization and three-dimensional reconstruction. Chromosoma. 1988;96:397–410. doi: 10.1007/BF00303033. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. . Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Fung JC, Sedat JW. Deconstructing the nucleus: global architecture from local interactions. Curr Opin Genet Dev. 1997;7:259–263. doi: 10.1016/s0959-437x(97)80136-0. [DOI] [PubMed] [Google Scholar]
- Mathog D, Hochstrasser M, Gruenbaum Y, Saumweber H, Sedat J. Characteristic folding pattern of polytene chromosomes in Drosophilasalivary gland nuclei. Nature. 1984;308:414–421. doi: 10.1038/308414a0. [DOI] [PubMed] [Google Scholar]
- Moroi Y, Hartman AL, Nakane PK, Tan EM. Distribution of kinetochore (centromere) antigen in mammalian cell nuclei. J Cell Biol. 1981;90:254–259. doi: 10.1083/jcb.90.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosgöller W, Leitch AR, Brown JKM, Heslop-Harrison JS. Chromosome arrangements in human fibroblasts at mitosis. Hum Genet. 1991;88:27–33. doi: 10.1007/BF00204924. [DOI] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, Pillus L, Gasser SM. The positioning of yeast telomeres depends on SIR3, SIR4, and the integrity of the nuclear membrane. Cold Spring Harb Symp Quant Biol. 1993;58:733–746. doi: 10.1101/sqb.1993.058.01.081. [DOI] [PubMed] [Google Scholar]
- Rabl C. Über Zelltheilung. Morphol Jahrb. 1885;10:214–330. [Google Scholar]
- Rae PMM, Franke WW. The interphase distribution of satellite DNA-containing heterochromatin in mouse nuclei. Chromosoma. 1972;39:443–456. doi: 10.1007/BF00326177. [DOI] [PubMed] [Google Scholar]
- Roth R, Halvorson HO. Sporulation of yeast harvested during logarithmic growth. J Bacteriol. 1969;98:831–832. doi: 10.1128/jb.98.2.831-832.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Loidl J, Schuster T, Schweizer D. Meiotic chromosome condensation and pairing in Saccharomyces cerevisiaestudied by chromosome painting. Chromosoma. 1992;101:590–595. doi: 10.1007/BF00360535. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Bähler J, Kohli J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J Cell Biol. 1994;127:273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Weich S, Schwegler H, Härle M, Heyting C, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selig S, Okumura K, Ward DC, Cedar H. Delineation of replication time zones by fluorescence in situ hybridization. EMBO (Eur Mol Biol Organ) J. 1992;11:1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling K, Lüdtke E-K. Arrangement of prematurely condensed chromosomes in cultured cells and lymphocytes of the Indian muntjac. Chromosoma. 1981;83:541–553. doi: 10.1007/BF00328278. [DOI] [PubMed] [Google Scholar]
- Sprague GF., Jr Assay of yeast mating reaction. Methods Enzymol. 1991;194:77–93. doi: 10.1016/0076-6879(91)94008-z. [DOI] [PubMed] [Google Scholar]
- Stack SM, Brown WV. Somatic pairing, reduction and recombination: an evolutionary hypothesis of meiosis. Nature. 1969;222:1275–1276. doi: 10.1038/2221275a0. [DOI] [PubMed] [Google Scholar]
- Steinmüller J, Schleiermacher E, Scherthan H. Direct detection of repetitive, whole chromosome paint and telomere DNA probes by immunogold electron microscopy. Chromosome Res. 1993;1:45–51. doi: 10.1007/BF00710606. [DOI] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjöld M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- Therman E, Denniston C. Random arrangement of chromosomes in Uvularia (Liliaceae) . Plant Syst Evol. 1984;147:289–297. [Google Scholar]
- Vourc'h C, Taruscio D, Boyle AL, Ward DC. Cell cycle-dependent distribution of telomeres, centromeres, and chromosome-specific subsatellite domains in the interphase nucleus of mouse lymphocytes. Exp Cell Res. 1993;205:142–151. doi: 10.1006/excr.1993.1068. [DOI] [PubMed] [Google Scholar]
- Weimer R, Haaf T, Krüger J, Poot M, Schmid M. Characterization of centromere arrangements and test for random distribution in Go, G1, S, G2, G1′, and early S′ phase in human lymphocytes. Hum Genet. 1992;88:673–682. doi: 10.1007/BF02265296. [DOI] [PubMed] [Google Scholar]
- Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- Zalensky AO, Allen MJ, Kobayashi A, Zalenskaya IA, Balhorn R, Bradbury EM. Well-defined genome architecture in the human sperm nucleus. Chromosoma. 1995;103:577–590. doi: 10.1007/BF00357684. [DOI] [PubMed] [Google Scholar]