Abstract
During mitosis, the ribbon of the Golgi apparatus is transformed into dispersed tubulo-vesicular membranes, proposed to facilitate stochastic inheritance of this low copy number organelle at cytokinesis. Here, we have analyzed the mitotic disassembly of the Golgi apparatus in living cells and provide evidence that inheritance is accomplished through an ordered partitioning mechanism. Using a Sar1p dominant inhibitor of cargo exit from the endoplasmic reticulum (ER), we found that the disassembly of the Golgi observed during mitosis or microtubule disruption did not appear to involve retrograde transport of Golgi residents to the ER and subsequent reorganization of Golgi membrane fragments at ER exit sites, as has been suggested. Instead, direct visualization of a green fluorescent protein (GFP)-tagged Golgi resident through mitosis showed that the Golgi ribbon slowly reorganized into 1–3-μm fragments during G2/early prophase. A second stage of fragmentation occurred coincident with nuclear envelope breakdown and was accompanied by the bulk of mitotic Golgi redistribution. By metaphase, mitotic Golgi dynamics appeared to cease. Surprisingly, the disassembly of mitotic Golgi fragments was not a random event, but involved the reorganization of mitotic Golgi by microtubules, suggesting that analogous to chromosomes, the Golgi apparatus uses the mitotic spindle to ensure more accurate partitioning during cytokinesis.
There are several inheritance strategies used by cellular organelles to facilitate equal partitioning into nascent daughter cells during cell division. At one extreme is the stochastic strategy, based on the laws of probability (Birky, 1983; Warren and Wickner, 1996). This is particularly suited to those organelles present in multiple, dispersed copies. The accuracy of partitioning depends on the number of copies (the more the better) and their distribution (the more evenly distributed the better). At the other extreme is the ordered partitioning strategy that is based on the mitotic spindle (McIntosh and Koonce, 1989). This ensures highly accurate segregation of chromosomes by tethering each daughter chromatid to one or the other of the mitotic spindle poles through microtubules attached to the kinetochores.
Membrane-bound organelles appear to use both strategies depending on the organism in which they find themselves. Thousands of randomly dispersed mitochondria are present in mammalian cells and are likely to be partitioned stochastically (Warren, 1993). However, mitochondria in the scorpion Centrurus attach themselves to the mitotic spindle poles and are partitioned along with the chromosomes (Wilson, 1916).
Low copy number organelles pose a particular problem for the inheritance process. In most, if not all animal cells, both the nuclear envelope/ER and Golgi apparatus exist as single or low copy organelles; at the onset of mitosis, both organelles fragment (Lucocq and Warren, 1987; Porter and Machado, 1960). Membranes of the nuclear envelope vesiculate in response to the disassembly of nuclear matrix proteins (Gerace and Blobel, 1980), and the ER undergoes variable degrees of fragmentation depending on the cell type. For the Golgi apparatus, fragmentation of the ribbon initially involves the release of the subunit stacks, each of which then undergoes extensive fragmentation to yield mitotic Golgi clusters (Lucocq et al., 1987). A hundred or more Golgi clusters are generated and thousands of vesicles, though the extent to which these remain attached to the clusters is still a matter of some debate. Since the Golgi apparatus in interphase cells is a compact reticulum in the juxtanuclear and often pericentriolar region of the cell, the generation of multiple fragments and their dispersal suggested a stochastic mode of inheritance.
Doubt has been cast on this suggestion after the recent characterization of mitotic Golgi clusters, which provided initial evidence that the partitioning of the Golgi apparatus was more accurate than would be expected for a stochastic process (Shima et al., 1997). However, the reason for this enhanced accuracy was not apparent, and although significant progress has been made using models that mimic mitotic alterations to Golgi membrane structure, the mechanisms responsible for the dispersal and partitioning of the Golgi ribbon in vivo remain obscure and controversial.
Here we have set out to analyze mitotic Golgi disassembly in living cells in hopes of understanding the nature of the partitioning mechanism. Two general models have been proposed to explain Golgi disassembly during mitosis. The first is based on inferences from ultrastructural studies and data from cell-free systems and suggests that during mitosis the Golgi apparatus fragments in situ to produce a heterogeneous collection of membranes that diffuse, resulting in the random distribution of membranes throughout the cell (Warren and Wickner, 1996).
The second model draws a parallel between the mechanisms of mitotic Golgi disassembly, and the fragmentation and redistribution of Golgi membranes after nocodazole-induced microtubule disruption. Based on recent work, it has been suggested that Golgi membrane dispersal after microtubule disruption, and during mitosis, could occur via retrograde transport of Golgi residents to the ER, and the subsequent reorganization of Golgi membrane fragments at ER exit sites (Cole et al., 1996; Lippincott-Schwartz and Smith, 1997). Since the ER is extremely well-dispersed throughout the cell, such a pathway would appear to effectively distribute Golgi residents and could presumably facilitate a passive inheritance of Golgi membranes based on the cytokinetic mechanism.
Here we report that contrary to previous expectations the Golgi is not partitioned via the ER, nor through a random distribution process, but rather find a surprising role for the mitotic spindle in the organization of Golgi membranes during mitosis.
Materials and Methods
Cell Culture and Microinjection
HeLa and Vero cells were cultured as previously described (Pepperkok et al., 1993; Shima et al., 1997). L929 fibroblasts and PtK1 cells were cultured in DME/high glucose (GIBCO BRL, Gaithersburg, MD) supplemented with 10% FCS (Sigma Chemical Co., St. Louis, MO), penicillin/ streptomycin (100 μg/ml), and 2 mM glutamine. Cell lines (HeLa, PtK1) harboring a chimera consisting of an enhanced green fluorescent protein (GFP)1 fused to the retention domain of N-acetylglucosaminyl-transferase were generated as previously described and all experiments with stable glycosyltransferase NAGTI (NAGFP) cell lines were performed after pretreatment with sodium butyrate to enhance transgene expression (Gorman and Howard, 1983; Shima et al., 1997).
Microinjection was performed using an automated microinjection system (Zeiss AIS; Carl Zeiss, Thornwood, NY), as previously described (Pepperkok et al., 1993). 2 mg/ml of cascade blue conjugated BSA was mixed with the DNA or protein as a coinjection marker. cDNAs encoding wild-type CHO Sar1p and a GTP-restricted mutant (H79G; here termed mSar1p) were provided by W.E. Balch (Scripps Research Institute, La Jolla, CA) and have been previously described (Aridor et al., 1995; Rowe and Balch, 1995). For cytoplasmic microinjections, a his-tagged version of mSar1p was isolated from bacteria using nickel affinity chromatography and size exclusion chromatography as described (Rowe and Balch, 1995).
To monitor the ER exit of newly synthesized proteins, mammalian expression plasmids encoding the plasma membrane marker CD8 (Nilsson et al., 1989) and the Golgi marker NAGFP were directly injected into nuclei, followed by a 45-min incubation at 37°C and the addition of cycloheximide (100 μg/ml) to inhibit further protein synthesis. Using this protocol, newly synthesized Golgi and plasma membrane markers could be detected at 60–80 min after injection.
For mitosis studies, prophase Hela cells were enriched by an aphidicolin (2.5 μg/ml) double block and release protocol (Shima et al., 1997). Brefeldin A (BFA; Boehringer Mannheim Corp., Indianapolis, IN) was used at 5 μg/ml.
Antibodies and Fluorescence Labeling
Cells on coverslips were either directly fixed in −20°C MeOH, or washed with Ca2+- and Mg2+-free PBS at 37°C, and preextracted for 1 min in PHEM buffer (60 mM PIPES, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.9) with 0.2% Triton X-100 and 10 μg/ml taxol to help preserve and visualize microtubules (Zhai et al., 1996). Samples were incubated in the corresponding antibodies, followed by 200 ng/ml Hoechst 33342 to visualize DNA, washed in PBS, and mounted in Mowiol.
Antibodies for these experiments include: affinity-purified rabbit antisera to rat GM130 (Nakamura et al., 1995); affinity-purified rabbit antisera to human giantin (Seelig et al., 1994); antisera recognizing the ER marker p62 (Mundy and Warren, 1992); monoclonal anti-ERGIC 53 (Schweizer et al., 1990); monoclonal anti–α-tubulin (clone B512) from Sigma Chemical Co.; monoclonal GTL2 raised against rat β-1,4 galactosyltransferase (Kawano et al., 1994). Rhodamine and fluorescein-conjugated secondary antibodies from Tago Inc. (Burlingame, CA); for microtubule labeling, Texas red X–conjugated secondary antibodies were obtained from Molecular Probes Inc. (Eugene, OR). Mitochondria were stained with nanomolar concentrations of MitoTracker Green FM (Molecular Probes Inc.) following the manufacturer's instructions.
Fluorescence Microscopy
Cells for live mitosis experiments were handled as described (Shima et al., 1997). Microinjection and live cell experiments were performed on a Zeiss Axiovert 135TV inverted microscope equipped with a ×63 Plan-Apochromatic oil objective. For live cells, a density filter (OD = 0.7%) was used to reduce light exposure to a minimum, exposure time was 0.2–0.4 s, and images were collected at 1-min or 5–10-s intervals. Images were recorded as previously described (Scales et al., 1997).
Analyses of nocodazole effects on mitotic Golgi were performed in the presence of dihydroethidium (0.5 μg/ml; Molecular Probes Inc.) to visualize DNA, and cycloheximide to avoid synthesis of new fluorescent protein. Phase microscopy and screening of stained nuclei were used to identify prometaphase cells. Nocodazole (10 μM; Sigma Chemical Co.) was added and an image of DNA and a z-series of the mitotic Golgi were obtained every 2–5 min using a laser scanning confocal microscope (MRC-1000; Bio-Rad Laboratories, Hercules, CA) as previously described (Shima et al., 1997). Images from the double label analysis of organelles and microtubules were also obtained using the same confocal system. All confocal images presented were two-dimensional projections of a z-series through the cell depth. Images were processed for presentation with Adobe Photoshop 3.0 (Adobe Systems, Mountain View, CA).
Results
Golgi Residents Do Not Cycle Through the ER during Mitotic Golgi Disassembly
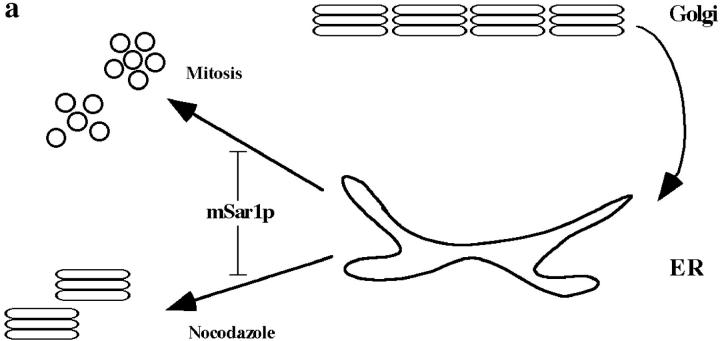
To test whether Golgi membranes were redistributed to the ER via retrograde transport during mitosis, we used a dominant mutant of the COPII vesicle coat GTPase Sar1p (H79G, here termed mSar1p), which has previously been shown to inhibit ER-to-Golgi transport (Barlowe et al., 1994; Kuge et al., 1994; Aridor et al., 1995). Microinjection of this mutant protein should result in the inhibition of ER export without affecting retrograde transport to the ER. Thus, under these conditions, any proteins that are redistributed from the Golgi region to the ER during mitotic disassembly should become trapped in the ER (Fig. 1 a).
Figure 1.
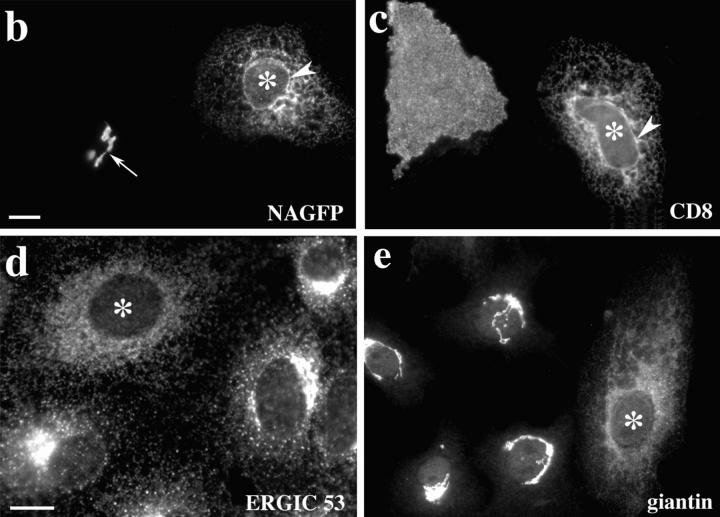
Transport inhibition by a dominant mutant of Sar1p (mSar1p). (a) Schematic representation of the Golgi-to-ER recycling model for Golgi disassembly, and the experimental approach using mSar1p to test this model. (b–e) Vero cells were microinjected with 2 mg/ml purified mSar1p mixed with 2 mg/ml cascade blue-conjugated BSA which served as a coinjection marker (not shown). After incubation of cells for 20 min at 37°C, they were injected a second time with plasmids encoding the plasma membrane marker CD8 or NAGFP and incubated as described in experimental procedures. (b) NAGFP-specific fluorescence; (c) staining for CD8; (d) staining for endogenous ERGIC 53. In cells injected with mSar1p (asterisks) all markers analyzed accumulated in the ER. In noninjected cells CD8 and NAGFP were transported to the plasma membrane or Golgi complex (marked by an arrow in b), respectively, and ERGIC 53 showed a typical distribution of the intermediate compartment. Arrowheads in b and c point to the nuclear envelope staining which is characteristic for ER localization. In e, mSar1p-injected (asterisk) and control cells were treated with 5 μg/ml BFA for 20 min, the BFA was washed out and cells fixed 2 h later and stained for the endogenous Golgi marker, giantin. In the presence of mSar1p, exit from the ER was inhibited for the Golgi resident giantin. Bars: (b, c, and e) 15 μm; (d) 10 μm.
We first looked in interphase cells at the effects of mSar1p on the ER exit of newly synthesized secretory proteins and on the distribution of a protein that normally recycles between the Golgi and ER. Microinjection of a purified recombinant version of mSar1p blocked ER export of newly synthesized Golgi residents (Fig. 1 b) and the plasma membrane marker CD8 (Fig. 1 c). Furthermore, endogenous ERGIC 53, a protein known to recycle between the Golgi and ER (Hauri and Schweizer, 1992), accumulated in an ER-like pattern (Fig. 1 d), and colocalized with the ER marker calnexin (not shown), suggesting it had recycled to the ER but could not escape from this compartment in the presence of the mutant protein.
Similar results were obtained injecting a plasmid encoding mSar1p instead of the purified recombinant protein, and injection of Sar1p wild type had no effect on ER to Golgi transport or recycling of ERGIC 53 (not shown, see also Aridor et al., 1995) excluding the possibility that the observed block in ER transport was a nonspecific effect of introducing increased amounts of a small GTPase into the cell cytoplasm.
Importantly, we also found that microinjection of mSar1p did not block the retrograde transport of Golgi residents to the ER after addition of the fungal metabolite BFA (Lippincott-Schwartz et al., 1989), and, in contrast to control-injected cells, the ER-localized Golgi residents in mSar1p-injected cells failed to exit the ER after BFA removal (Fig. 1 e).
From these results, we concluded that mSar1p was a suitable tool to examine whether Golgi residents dispersed via the ER during mitotic disassembly. If true, recycling Golgi residents should accumulate in the ER in the presence of mSar1p and block the formation of mitotic Golgi fragments.
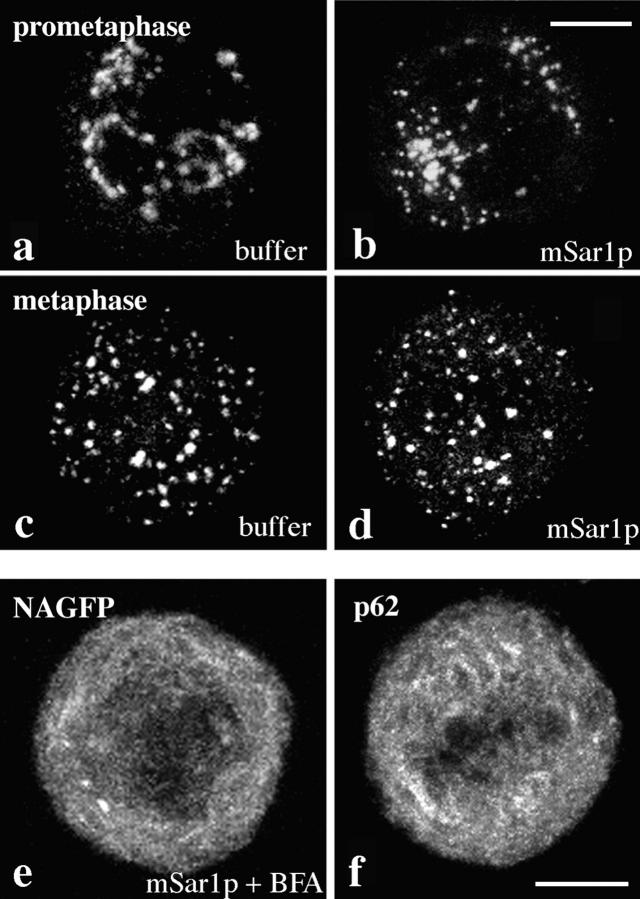
Cells expressing a previously characterized Golgi marker consisting of GFP and the Golgi retention domain of the NAGFP (Shima et al., 1997), were synchronized in G1/S phase of the cell cycle by a double aphidicolin block. Removal of the drug resulted in an increase of G2/M-phase cells with a maximum between 10 and 12 h after release of the block (Shima et al., 1997). Cells that had not entered mitosis at 9 h after release of the aphidicolin block were microinjected with mSar1p. Numerous mitotic cells were observed 1–2 h after injection, suggesting that mSar1p did not interfere with entry into mitosis. Most importantly, the Golgi markers NAGFP and giantin (not shown) did not appear to accumulate in the ER at any time after microinjection (n = 20 cells), and mitotic Golgi fragmentation proceeded in a manner indistinguishable from buffer- injected cells (Fig. 2). The appearance and distribution of Golgi fragments in mSar1p-injected metaphase cells (Fig. 2 d) was similar to Golgi fragments in buffer-injected metaphase cells (Fig. 2 c), and the presence of mSar1p did not inhibit the ability of Golgi residents to fuse with the ER in the presence of BFA (Fig. 2 e). For comparison, the appearance of the ER in mitosis is shown (Fig. 2 f).
Figure 2.
Analysis of mitotic Golgi fragmentation in mSar1p injected cells. HeLa cells stably expressing NAGFP were enriched in late G2, and cells that had visibly not entered mitosis were microinjected with mSar1p and cascade blue conjugated BSA (b and d) or with cascade blue conjugated BSA only (a and c). 1 h after injection cells were fixed and mitotic cells analyzed by confocal microscopy. Projections of 10–25 z-sections (0.5-μm steps) ranging from the bottom to the top of the cells are shown. In e, cells were treated with 2 μg/ml BFA at the time of injection; in f staining for p62, an ER membrane protein is shown. Bar, 10 μm.
Taken together, these results do not support a Golgi fragmentation model that relies on the recycling of Golgi residents through the ER.
Direct Visualization of Golgi Fragmentation during the Prophase to Metaphase Transition in Living Cells
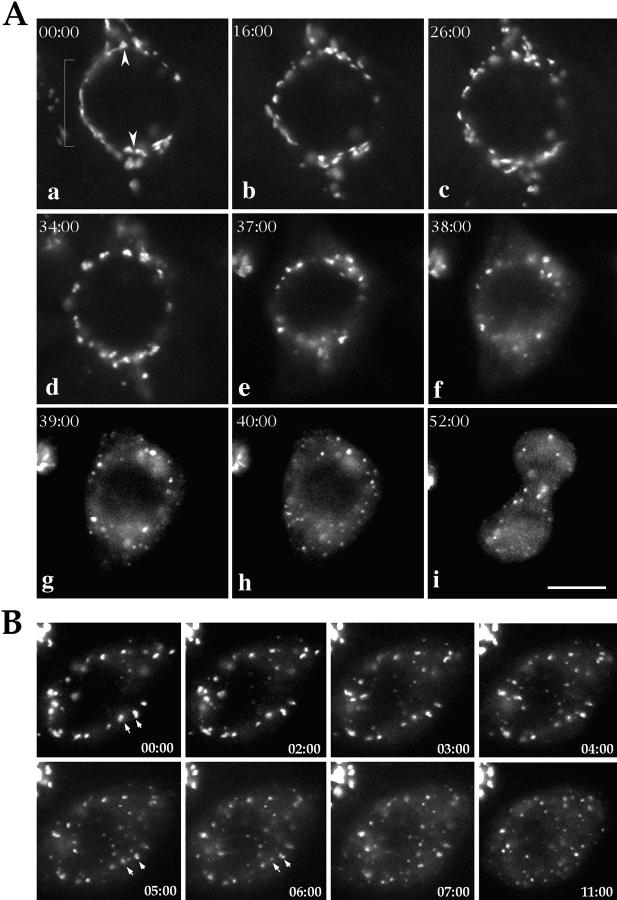
To further clarify the mode of Golgi disassembly during mitosis, we studied mitotic Golgi breakdown by directly visualizing Golgi membranes in synchronized, living HeLa cells using the NAGFP cell line (Shima et al., 1997). Early during prophase the interphase Golgi ribbon was altered, now extending from its original polarized position to surround much of the cell nucleus (Fig. 3 a), with a distinct organization of Golgi material in regions destined to become the cell poles (Fig. 3 a, arrowheads). As prophase progressed the Golgi ribbon fragmented, but there was little alteration to membrane distribution around the cell nucleus. This first stage of Golgi breakdown lasted between 30–45 min (Fig. 3, a–d) and using confocal microscopy it was estimated that the Golgi apparatus at this stage exists as 35–45 fragments with a diameter of 1–3 μm (long axis).
Figure 3.
Direct visualization of Golgi membranes during mitosis. (A) Images were acquired every 5 min to see changes in Golgi morphology indicating that cells were in late G2 or the beginning of prophase. Thereafter images were acquired every minute, and cells were followed through all phases of mitosis. Images in a–d show early prophase cells before the breakdown of the nuclear envelope. In e–h the prophase to metaphase transition is shown, and i shows the same cell in telophase. Arrows in a point to Golgi fragments organized around the position of the emerging spindle poles. (B) A series of images (1 min interval) acquired at the prophase to metaphase transition are shown. 02:00 marks the beginning of the second stage of fragmentation and a period of extensive Golgi scattering. Mitotic Golgi fragments are designated by arrows. A quicktime movie comprising the images acquired will be available on the internet. Bar, 15 μm.
Coincident with the rounding of mitotic cells and the apparent breakdown of the nuclear envelope was a second, much shorter stage of fragmentation that was characterized by numerous divisions of large Golgi fragments, followed by their redistribution throughout the cell over a 2–5-min period as the cell progressed into metaphase (Fig. 3, e–h).
The dynamics of Golgi membranes during these two stages of fragmentation were analyzed in greater detail (Figs. 3 and 4). In the first stage, the mitotic Golgi appeared to fragment in situ, with little if any diffusion-mediated displacement, suggesting that despite breakdown of the ribbon, Golgi membranes remain tethered to an underlying structure (Fig. 3 a, bracket denotes region of particular interest).
Figure 4.
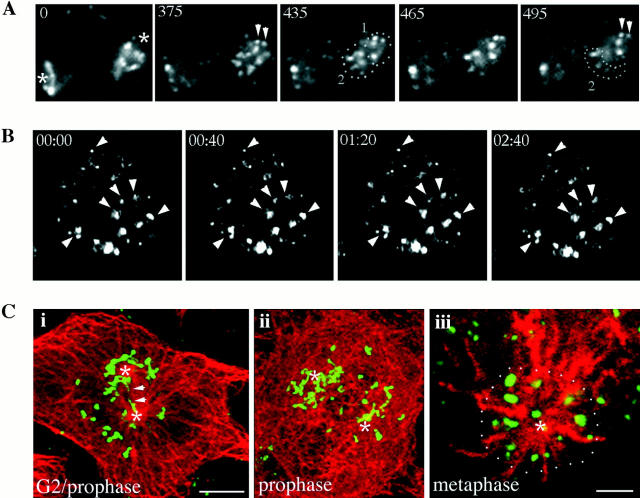
Analysis of HeLa cell mitotic Golgi fragment dynamics and subcellular localization. (A) Images were acquired every 10 s to track Golgi fragment movements in the future polar regions during the prophase to metaphase transition. Asterisks in 0 indicate Golgi fragments associated with the spindle poles (identified by phase microscopy and subsequent location of metaphase chromosomes). Dotted lines indicate the emergence of a stable, non-mobile population of Golgi (group 1), and a population that reorganizes and disperses during the period of observation (group 2). Arrowheads indicate Golgi fragments stably associated with the spindle poles during the entire period of observation. (B) Dynamics of Golgi fragments in metaphase cells were analyzed by confocal microscopy acquiring five z-sections every 20 s. Shown is a two-dimensional projection of the z-series of images. Metaphase Golgi fragments are stable (see arrowheads) and undergo only insignificant redistribution. (C) Double label analysis of the Golgi marker GM130 (green) and microtubules (red) in HeLa cells. (i) Golgi membranes associate with microtubules of the separating centrosomes (asterisks). Arrows designate membranes that appear to be distributing between the two separating centrosomes. (ii) By late prophase Golgi membranes are situated around the developing asters on either side of the nucleus. (iii) An en face view of the long axis of the metaphase spindle pole demonstrates the organization of a subset of Golgi membranes in a circular array surrounding the aster. Bar: (i and ii) 5 μm; (iii) 1 μm.
The second stage of fragmentation occurred during the prophase to metaphase transition and was characterized by at least two dispersal steps. The first was a centripetal movement of perinuclear Golgi fragments in parallel with the apparent collapse in the nuclear envelope as cells rounded (Fig. 3, compare d with e). This was followed by the scattering of fragments throughout the cytoplasm of the rounding cell (Fig. 3 A, f–h and B).
Next, we examined the second stage of fragmentation at higher time resolution (one image every 10 s), focusing specifically on the Golgi fragments concentrated in the putative polar regions (determined by phase appearance and marked by asterisks in Fig. 4 A, t = 0 s). This revealed a gradual rearrangement of mitotic Golgi fragments. The example shown in Fig. 4 shows how in the course of ∼2 min, a collection of polar fragments becomes reorganized into two subpopulations: one that remains in a relatively stable configuration in a region proximal to the developing spindle pole (Fig. 4 A, designated population 1) and one that moves away from the pole region (Fig. 4 A, designated population 2). The average speed of mitotic fragments during the prophase to metaphase transition was calculated to be 1–3 μm/min. Saltatory movements, indicative of microtubule motor-based movements, were not observed.
In contrast to the dynamic reorganization of Golgi membranes during the prophase-to-metaphase transition, in metaphase, the fragmentation and dispersal process ceased (Fig. 4 B). Live cell confocal microscopy showed that the majority of mitotic Golgi fragments maintained their positions relative to each other throughout the period of observation (∼3 min; see arrows). This behavior is in accord with previous work that suggested that the relative positions of mitotic Golgi were fairly constant throughout the metaphase to telophase period (Shima et al., 1997).
Taken together, these data suggested that the fragmentation of mitotic Golgi was achieved in two stages, the second of which involved extensive scattering of the Golgi membranes. This scattering was coincident with the major structural rearrangements of the nucleus and microtubule cytoskeleton (Zhai et al., 1996) and involved the gradual, regulated redistribution of some Golgi fragments, while others appeared to be stably situated around the developing mitotic spindle poles. These observations suggested to us that Golgi behavior may be regulated by microtubules during mitosis.
Organization of Mitotic Golgi Fragments by the Emerging Mitotic Spindle
The appearance of microtubules and mitotic Golgi fragments in fixed HeLa cells was consistent with the idea that the fragmented Golgi membrane population segregated into two groups, each in apparent association with microtubules from one of the centrosomes or spindle poles (Fig. 4 C). In early prophase, Golgi membranes appeared to be distributing between the two separating centrosomes (Fig. 4 C, i, see arrows), and by late prophase appeared to attain a loose distribution on either side of the future division plane (Fig. 4 C, ii, asterisks mark poles). Importantly, similar to the findings for the progression from prophase to metaphase in living cells (Fig. 4 A), after the scattering process, a radial array of Golgi fragments remained associated with the spindle pole (Fig. 4 C, iii).
However, the variability in the position of the spindle and the extensive degree of out-of-focus material in the round HeLa cells precluded a thorough assessment of the spatial relationship of the Golgi and the extensive mitotic microtubule cytoskeleton. To overcome this limitation, we decided to continue morphological analysis in other cell lines such as PtK1, which stay attached to the substratum during mitosis, and therefore have a flattened cell morphology and provide a superior view of both the spindle apparatus and cytoplasm during mitosis (Rieder et al., 1994).
In early prophase PtK1 cells, the Golgi ribbon appeared disrupted and fragments were detected on either side of the nuclear envelope (compare Fig. 5, a with b). By prometaphase (Fig. 5 c), the majority of mitotic Golgi membranes were organized radially around the developing spindle asters (marked by asterisks). As observed in HeLa cells, the radial array of Golgi fragments separated into two populations at metaphase: one, a concentration of membranes around the spindle pole (Fig. 5 d, asterisks), the other, dispersed in the periphery and often situated in close proximity to microtubules (Fig. 5 d, arrows). This organization of Golgi clusters around the spindle poles and associated microtubules persisted through anaphase (Fig. 5 e) and telophase (Fig. 5 f). Further support for the organizing influence of the mitotic spindle on Golgi fragments was seen in multispindle cells (Fig. 5 g), which show a concentration of mitotic Golgi membranes surrounding each of the spindle poles (arrows). In contrast, mitochondria (Fig. 5 h) and ER (data not shown and Ioshii et al., 1995) demonstrated no such organization of membranes at the spindle poles, suggesting that the mitotic spindle does not have an organizing influence on all membrane-bound organelles. Similar organization of mitotic Golgi was also observed in L929 fibroblasts, Vero cells and PtK2 cells (data not shown).
Figure 5.
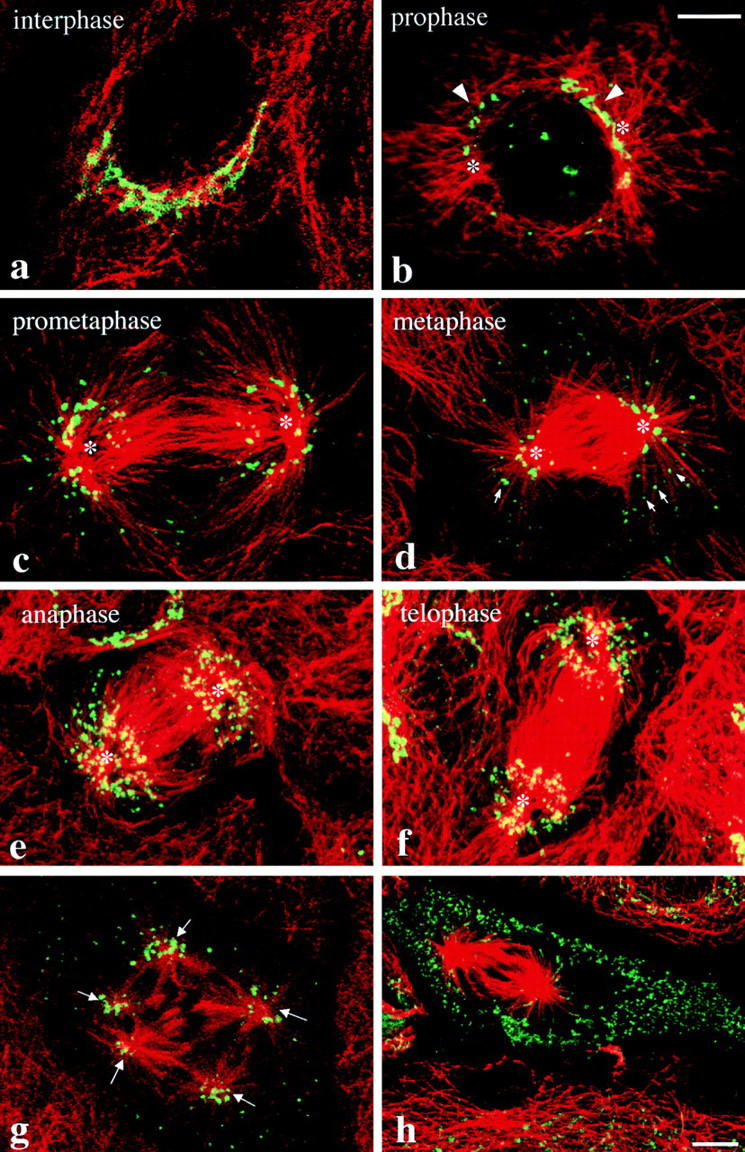
Appearance of Golgi membranes and microtubules during mitosis in PtK1 cells. Exponentially growing PtK1 cells were fixed and stained for the Golgi marker GM130 (green, a–g) or mitochondria (see Material and Methods; green, h) and microtubules (red). The cells shown in different images represent different phases of mitosis characterized by their organization of microtubules/ chromatin. Asterisks indicate the centrosomal/spindle pole regions. Arrowheads (b) designate Golgi fragments that were frequently detected on either side of the nuclear envelope by the earliest indications of entrance into mitosis. Arrows indicate Golgi fragments in proximity to aster microtubules (d), or concentrated around spindle poles (g). Bar: (a–g) 10 μm; (h), 5 μm.
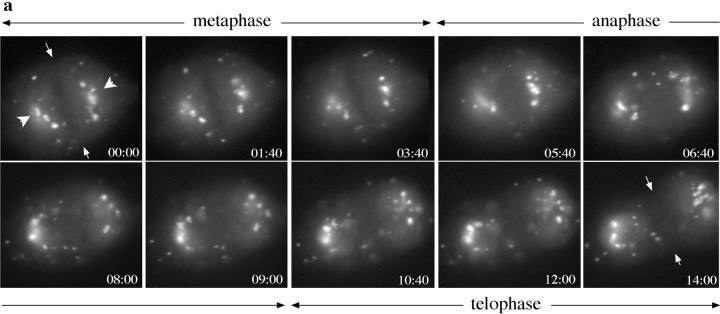
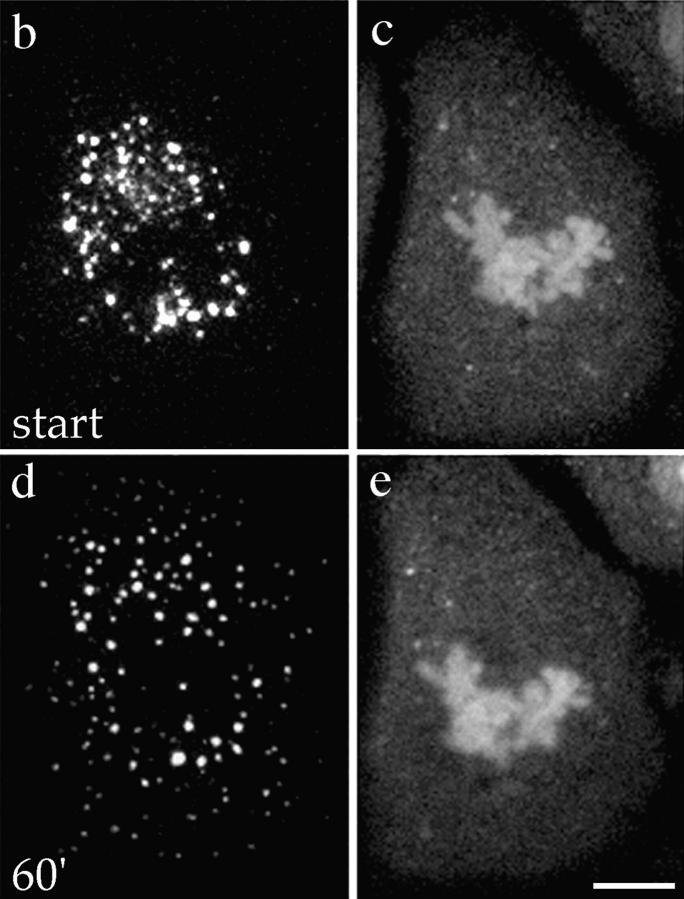
These data suggest that the distribution of mitotic Golgi clusters correlates with the position of the mitotic spindle. Further support for the role of spindle microtubules in mitotic Golgi organization was provided by analysis of the behavior of mitotic Golgi fragments in living mitotic PtK1 cells expressing the NAGFP Golgi marker. Living metaphase PtK1 cells exhibited a concentration of Golgi membranes on either side of the metaphase plate, and were accompanied by a less abundant population of mitotic fragments that were apparent in the peripheral cytoplasm (Fig. 6 a). This distribution of mitotic Golgi was maintained throughout the major periods of chromosome/spindle pole movements (anaphase) and was accompanied by the poleward movement and eventual segregation of Golgi membranes into the emerging daughter cells during the initiation of the cleavage furrow (telophase). Treatment of prometaphase cells with nocodazole, which results in the preferential loss of the dynamic spindle microtubule population (Cassimeris et al., 1990; Zhai et al., 1996), resulted in the redistribution of mitotic Golgi membranes from the spindle region (Fig. 6, b and c) to the cell periphery (Fig. 6, d and e), further implicating microtubules in the mitotic organization of Golgi membranes.
Figure 6.
Organization of mitotic Golgi membranes by microtubules in living PtK1 cells. (a) A series of images acquired during the metaphase-to-telophase progression in a PtK1 cell line stably expressing the NAGFP chimera. Images were taken at 20-s intervals. Still images highlight the concentration of mitotic Golgi (arrowheads) on either side of the metaphase plate (arrows in first panel), presumably in association with elements of the mitotic spindle. This peri-spindle distribution of mitotic Golgi was maintained while Golgi membranes were partitioned into the daughter cells. A quicktime movie comprising the images acquired will be available on the internet. (b–e) A living prometaphase cell stably expressing NAGFP (b and d) was stained with dihydroethidium (c and e) to label DNA as described in Materials and Methods, and visualized using confocal microscopy. 10 z-sections through the entire depth of the cell were taken either before or at different time-points (shown is 60 min) after the addition of nocodazole (10 μM) and cycloheximide (100 μg/ml). Two-dimensional projections of the z-sections are shown. After disruption of the mitotic spindle with nocodazole, the peri-spindle organization of Golgi clusters redistributes (d), with mitotic Golgi appearing more frequently at the cell periphery. Bar, 10 μm.
Reexamination of Golgi-to-ER Transport in Nocodazole
The experimental data in living cells provided evidence that mitotic disassembly of the Golgi occurs through membrane fragmentation and dispersal. Together with the results obtained using mSar1p in mitosis, it appears that Golgi fragmentation and dispersal can be accomplished without the need to invoke Golgi-to-ER retrograde transport, and may rely upon the integrity of microtubules. Thus, we wanted to determine if transit of Golgi residents to the ER was required to redistribute Golgi membranes after microtubule disruption by nocodazole, the experimental system that has led to the recent interest in the ER transit model for Golgi disassembly (Cole et al., 1996).
Therefore, we examined the effects of mSar1p on nocodazole-induced Golgi fragmentation. Cells were incubated for 30 min at 4°C, which depolymerizes cold-sensitive microtubules without disrupting the structure of the juxtanuclear Golgi ribbon at the light microscopic level (data not shown; Cole et al., 1996; Turner and Tartakoff, 1989). Purified recombinant mSar1p was injected into cells that were maintained in the cold in the presence of nocodazole, and subsequently the cells were incubated for different time points at 37°C in the continuous presence of nocodazole. All experiments also included the protein synthesis inhibitor cycloheximide to ensure that the analysis was restricted to recycling of proteins in the ER, rather than newly synthesized proteins.
In mSar1p-injected cells, ERGIC 53 accumulated in the ER (Fig. 7, a and c); however, Golgi membranes appeared to fragment and distribute throughout the cell with similar kinetics as noninjected cells (Fig. 7, b and d). At no time point (between 15 min and 3 h) after injection did Golgi markers appear in the ER. Similar results were obtained when cells were first treated with nocodazole for 2 h to disperse the Golgi and then injected with mSar1p (Fig. 7, e and f) demonstrating that ERGIC 53 cycles between Golgi fragments and the ER in the absence of microtubules. In contrast, Golgi residents were retained in the peripheral Golgi fragments, suggesting that, after their redistribution to the periphery, Golgi residents do not recycle through the ER.
Figure 7.
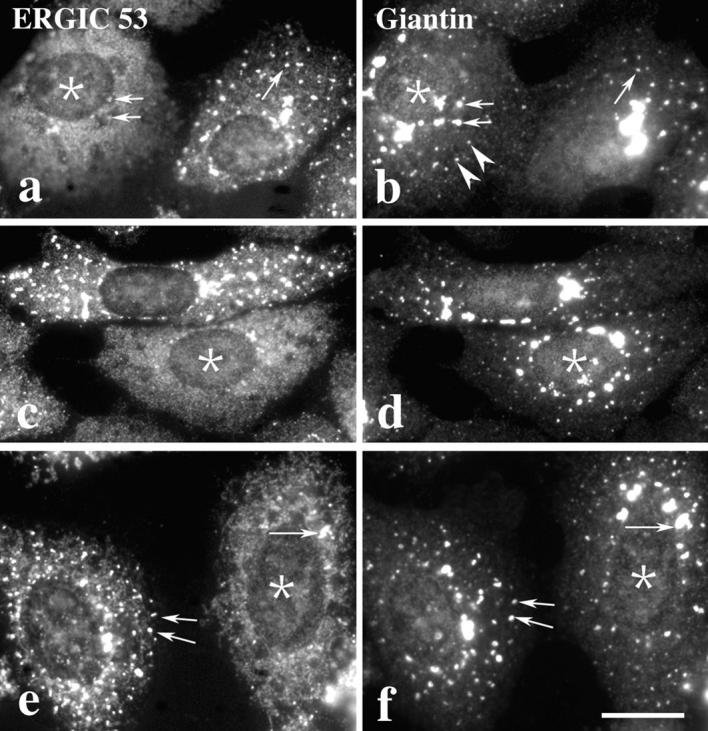
Analysis of nocodazole induced Golgi fragmentation in mSar1p injected cells. Vero cells were incubated for 30 min at 4°C to depolymerize microtubules. Cells were microinjected in the cold with 2 mg/ml purified mSar1p mixed with 2 mg/ml cascade blue conjugated BSA. Immediately after injection, cells were incubated for 1 h (a and b) or 2 h (c and d) at 37°C in the presence of nocodazole (10 μM) before they were fixed and stained for ERGIC 53 (a, c, and e) or the Golgi marker giantin (b, d, and f). In e and f, cells were first treated with nocodazole for 2 h, microinjected and subsequently incubated in the presence of nocodazole for 1 h. Microinjected cells (marked by asterisks) were identified by the coinjected BSA (not shown). Microinjection of mSar1p results in the accumulation of ERGIC 53 in the ER but does not interfere with the formation and distribution of the Golgi fragments (marked by arrowheads in b); in noninjected cells ERGIC 53 and giantin often appear to colocalize in peripheral Golgi fragments (marked by arrows). Bar, 15 μm.
Discussion
Ultrastructural observations laid the foundation for an initial understanding of Golgi behavior in animal cells during mitosis by describing the transformation of the Golgi ribbon into a dispersed, heterogeneous collection of tubular networks, short tubules, and vesicles (Lucocq et al., 1987). Cell-free analysis of Golgi disassembly/reassembly reinforced these earlier ultrastructural findings by providing a potential molecular basis for the process of mitotic vesiculation and subsequent reassembly of the Golgi apparatus (Misteli and Warren, 1994; Rabouille et al., 1995). These observations had given rise to the model that the pericentriolar, polarized Golgi was converted into numerous, well-distributed membranes to increase the accuracy of achieving equal partitioning solely through the act of dividing a cell into equal halves, an event termed stochastic partitioning.
However, recent analysis of Golgi membranes during the metaphase to telophase period demonstrated a precise regulation of their number and compartmentation, and suggested that the accuracy of partitioning the mitotic Golgi exceeded that expected for a stochastic partitioning process (Shima et al., 1997). Using a combination of microinjection and microscopic techniques, we have investigated two alternative models to explain the mitotic disassembly of the mammalian Golgi apparatus with the anticipation that these studies may shed light on the mode of mitotic Golgi partitioning. In summary, the data obtained reveal that neither of the two existing models is sufficient to describe the mitotic Golgi disassembly process. Instead, the direct visualization of Golgi behavior, together with the analysis of microtubule distribution by immunocytochemistry suggests that mitotic Golgi disassembly is a progressive fragmentation and dispersal process that is regulated by mitotic microtubules.
The Golgi–ER Retrograde Transport Model
Recent work on the fragmentation and redistribution of Golgi membranes after microtubule disruption (Cole et al., 1996) has led to the proposal that the mitotic disassembly of the Golgi apparatus may occur via retrograde transport of residents to the ER (Lippincott-Schwartz and Smith, 1997).
To directly address this model, we used a dominant interfering mutant of the COPII GTPase Sar1p (mSar1p) to devise a strategy that would result in the accumulation of Golgi components in the ER if they cycled through this compartment during either microtubule disruption or mitosis. Microinjection of mSar1p blocked ER export under various conditions but did not interfere with the transport of ERGIC 53 from the Golgi to the ER or the relocation of Golgi residents to the ER after treatment with BFA. In contrast to the predictions of the Golgi–ER retrograde transport model, microinjection of mSar1p during microtubule disruption by nocodazole did not result in an accumulation of Golgi residents in the ER, and redistribution of Golgi fragments to the periphery occurred with similar kinetics as in control cells. However, in the same cells ERGIC 53 accumulated in the ER demonstrating that retrograde transport of bona fide recycling proteins from the Golgi region to the ER was intact, but not ER export. Similarly, morphological examination of cells injected with mSar1p showed that they were able to undergo mitotic disassembly in a manner indistinguishable from control-injected cells, forming a typical fragmented and dispersed collection of membranes.
The most likely explanation for these results is that during mitosis or after microtubule disruption, Golgi residents redistribute via pathways that do not involve their recycling through the ER. However, an alternative explanation could be that during mitosis or microtubule disruption, Golgi residents recycle through the ER via a Sar1p-independent mechanism and therefore would escape detection in these experiments. Given the sum of current evidence suggesting that the COPII pathway is primarily responsible for packaging and budding of ER-derived vesicles (Kuge et al., 1994; Aridor et al., 1995; Barlowe, 1995), and the control experiments that showed that newly synthesized cargo molecules and Golgi residents, known recycling proteins, and BFA-relocated Golgi residents all require normal Sar1p function to exit the ER, this possibility seems unlikely.
Our experimental findings do not exclude the existence of a recycling pathway that could, under certain conditions, transport Golgi residents to the ER. For example, experiments using a Golgi-localized version of a VSV temperature-sensitive folding mutant (Cole et al., 1998), BFA (Lippincott-Schwartz et al., 1989), and overexpression of a mutant form of the rab6 GTPase (Martinez et al., 1997), suggest that a Golgi–ER retrograde transport pathway can at least be induced under certain conditions. However, it is still not clear how these experimental systems are related to physiological membrane traffic events.
The period of observation for our microtubule disruption experiments was restricted to a maximum of 3 h; therefore, we cannot exclude the possibility of recycling that occurs over lengthier time frames. However, if such a pathway exists, it is very unlikely to play a role in the process of Golgi disassembly after nocodazole treatment or during mitosis, which are essentially complete within 90– 120 min (Cole et al., 1996) and 30–50 min (see Fig. 3 and Misteli and Warren, 1995), respectively.
Mitotic Fragmentation of the Golgi Ribbon In Vivo
Since Golgi-to-ER recycling during mitosis appeared to be unlikely, we turned to investigate the possibility of a Golgi fragmentation and a random dispersal process by using the NAGFP–HeLa cell line to document the behavior of the Golgi during mitotic disassembly in living cells.
The first phase of mitotic disassembly lasted for 30–45 min and was characterized by an in situ transformation of the ribbon-like Golgi into fragments that were roughly situated around the nucleus. Because this phase of fragmentation occurred quite early during prophase, it is likely to correspond to the transformation of the interphase ribbon into dispersed Golgi stacks (Colman et al., 1985), which has been carefully examined at the ultrastructural level (Lucocq et al., 1987; Misteli and Warren, 1995). Virtually nothing is known of the molecular mechanisms responsible for the redistribution of stacks at this stage of mitosis; however, the similarity between this stage of disassembly and the effects of microtubule disruption on the Golgi ribbon raises the possibility that alterations in microtubule organization that accompany centrosome duplication/separation may play a role.
The second stage of disassembly was brief, occurring within a 5-min period, and was characterized by additional fragmentation and the bulk of the dispersal of the mitotic Golgi. These changes were coincident with the loss of the nuclear envelope and the rounding of cells, events that mark the prophase to metaphase transition. Ultrastructural observation has shown that the Golgi stacks are transformed into a heterogeneous collection of tubular networks, short tubules, and vesicles, termed mitotic clusters, during the transition from late prophase to metaphase (Lucocq et al., 1987); therefore, it is likely that the second phase of fragmentation represents the change from stacks to mitotic clusters. Abundant evidence supports a role for a peak in cdc2 kinase activity in triggering the structural changes to the nuclear envelope and microtubules during the prophase to metaphase transition (Simos and Georgatos, 1992; Jackman et al., 1995; Macaulay et al., 1995), therefore, raising the possibility of a similar role for cdc2 kinase in the second phase of Golgi disassembly. In further support of this idea, cdk-dependent phosphorylation of the Golgi matrix protein GM130 has recently been implicated as a critical step in the mitotic disassembly of Golgi membranes using a cell-free system (Nakamura et al., 1997).
Although the findings derived from live cell observation supported a model of mitotic Golgi disassembly based on progressive fragmentation, several features of mitotic Golgi behavior were clearly in contradiction to the concept that fragmentation facilitated the random distribution of Golgi membranes.
First, the major dynamic period for mitotic Golgi fragments is highly regulated, occurring within a well-defined temporal window just before metaphase, and terminating with the establishment of a well-dispersed, yet immobile collection of metaphase Golgi membranes. Furthermore, movement of individual mitotic Golgi fragments during the prophase to metaphase transition was not based on random diffusion. The dispersal process began with mitotic, perinuclear Golgi fragments moving in a centripetal manner, coordinately with the movement of the collapsing cell and nuclear membranes during cell rounding. Also, subsets of mitotic Golgi membranes remained immobile, while others appeared to relocate throughout the cell. Lastly, and most importantly, random distribution of Golgi membranes is not sufficient to explain their distinct organization that could be tracked to the vicinity of the emerging spindle poles. These results implied the existence of an underlying structure that was involved in regulating Golgi membrane position and dynamics during mitosis.
Organization of Golgi Fragments by the Mitotic Spindle
Analyses of microtubules and Golgi markers by confocal immunofluorescence microscopy were consistent with the live-cell data suggesting that early mitotic Golgi membranes were organized around the two pole regions of the emerging mitotic spindle. Furthermore, mitotic Golgi fragments frequently appeared associated with astral microtubules, both proximal and distal to the spindle, strongly suggesting a role for the microtubules themselves in mitotic Golgi organization. This hypothesis is supported by the observation that the organization of mitotic Golgi is disrupted after treatment of prometaphase cells with the microtubule disrupting drug nocodazole. However, additional work is required to determine if the role of microtubules is direct, or rather serves to organize another element of the cytoskeleton that directly regulates Golgi behavior during mitosis.
An interaction of mitotic Golgi with elements of the developing mitotic spindle could provide a potential explanation for our observations on the behavior of the Golgi during mitosis, such as the regulated tethering of Golgi membranes, and the organization of prophase Golgi in the future spindle region. Furthermore, the major period of mitotic Golgi dispersal, during the prophase-metaphase transition, is coincident with an abrupt depolymerization and rearrangement of microtubules to form the mitotic spindle (Zhai et al., 1996). Therefore, one speculative scenario is that the tethering, organization, and displacement processes are all accomplished by the association of mitotic Golgi with the reorganizing microtubules of the evolving spindle. The observation of live cells at higher time resolution suggested a gradual reorganization of mitotic Golgi fragments around the developing poles; a process that was incompatible with fragment diffusion, but was also slower (1–3 μm/min) than expected for motor protein-directed movement in the secretory pathway (0.5–3 μm/s; Lippincott-Schwartz et al., 1995; Scales et al., 1997), and lacked saltatory movements often associated with motor protein–driven displacement. The failure to observe clear indications of motor protein–dependent movement was consistent with previous reports showing that motor-dependent plus and minus end–directed movement of membranes on microtubules is inhibited during mitosis (Allan and Vale, 1991). Therefore, in the absence of obvious motor driven displacement, we suggest that the dynamics of the microtubules themselves may provide the force involved in reorganization of mitotic Golgi fragments. For example, microtubules of the developing half spindle are believed to exert an away from the pole pushing force (“polar wind”) on chromosomes that could potentially play a role in the reorganization of mitotic Golgi seen in the prometaphase spindle pole (Rieder and Salmon, 1994). Similarly, association with the growing and shrinking ends of microtubules is sufficient to translocate membranes in vitro, via a tip attachment complex (TAC), which may be a source for the tethering and reorganizing force exerted on Golgi membranes during mitosis (Waterman-Storer et al., 1995). Interestingly, recent work has suggested a potential role for microtubule shrinkage and/or growth in a slow class (<0.1 μm/s) of phagosome movements in vivo (Blocker et al., 1998).
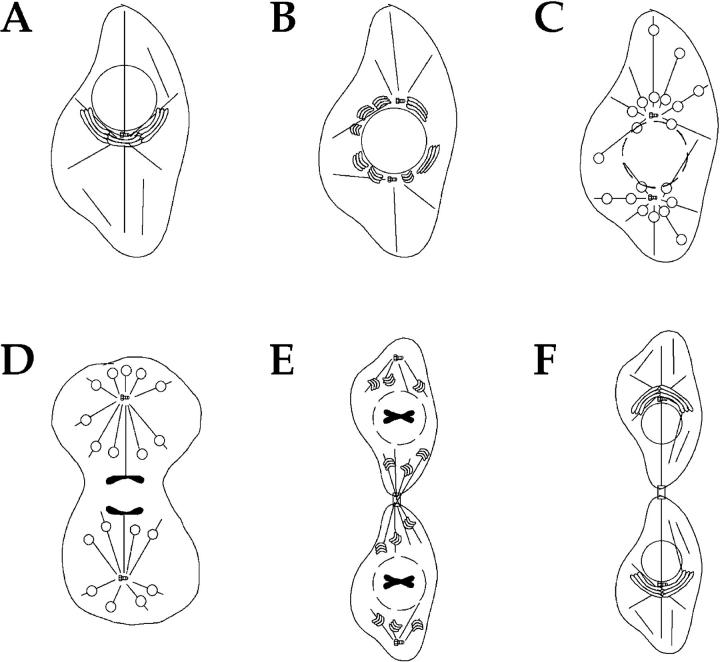
Through an analysis of mitotic Golgi disassembly in vivo we have been able to provide direct evidence for the existence of an ordered inheritance strategy for the Golgi apparatus during cell division. Moreover, the experimental findings raise several interesting issues concerning the behavior of Golgi membranes during mitosis. Foremost, why do Golgi membranes disassemble into dispersed tubulo-vesicular clusters during mitosis? The previous belief was that the fragmentation process facilitated a chance-based partitioning, but the organization of mitotic Golgi by microtubules casts doubt on this notion. Perhaps, instead, the disassembly process facilitates the allotment of the interphase Golgi ribbon into two populations, each organized (directly or indirectly) by the microtubules of a developing half-spindle (Fig. 8). In fact, the early segregation of Golgi membranes into two centrosome-associated populations (see Figs. 4 and 5) suggests that the partitioning process could potentially be initiated at the earliest stages of mitosis, in prophase, when the centrosomes separate and nucleate their astral arrays. This mode of partitioning shares general features with the organization and separation of chromosomes during cell division, a paradigm that should provide a useful framework to facilitate the future dissection of the inheritance mechanism for the Golgi apparatus.
Figure 8.
Model for Golgi apparatus disassembly and partitioning during mitosis. (A) In most cells, the Golgi apparatus exhibits a polarised position in interphase, at one side of the nucleus and next to the centrosome. (B) At the G2/M transition, the Golgi ribbon reorganizes and adopts a more perinuclear localization. This reorganization may be coordinated by the separation of centrosomes, and the association of Golgi membranes with the two microtubules organizing arrays. (C) Fragmentation into Golgi stacks continues throughout prophase, and coincident with nuclear envelope break down, Golgi stacks rapidly fragment to yield mitotic Golgi clusters that are repositioned around the spindle poles and by astral microtubules. (D) As cell division ensues, Golgi clusters associate with microtubules, forming ring-like structures that are partitioned along with each spindle pole and one complement of sister chromatids. (E) At the end of cytokinesis, reformed Golgi stacks are positioned both close to the midbody and to the daughter cell centrosome, then slowly converge to reform the Golgi ribbon as shown in F (Moskalewski and Thyberg, 1990).
Acknowledgments
We would like to thank W.E. Balch for supplying the Sar1p plasmids; M. Lowe for assistance in purification of Sar proteins; H.P. Hauri, D. Mundy, M. Renz, and T. Suganuma for supplying antibodies; A.J. Stokes and P. Jordan for technical assistance with microscopy; and J. Herreros, M. Lowe, and C. Ruhrberg for critical review of the manuscript.
Abbreviations used in this paper
- BFA
brefeldin A
- COP
coat protein
- ERGIC
endoplasmic reticulum–Golgi intermediate compartment
- GFP
green fluorescent protein
- NAGFP
glycosyltransferase NAGTI
- TAC
tip attachment complex
Footnotes
D.T. Shima is a Hitchings-Elion fellow supported by the Burroughs Wellcome Fund and N. Cabrera-Poch is a recipient of a Postdoctoral Fellowship from the Spanish Ministry of Education and Culture.
Drs. Shima and Cabrera-Poch contributed equally to this work.
References
- Allan VJ, Vale RD. Cell cycle control of microtubule-based membrane transport and tubule formation in vitro. J Cell Biol. 1991;113:347–359. doi: 10.1083/jcb.113.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M, Bannykh SI, Rowe T, Balch WE. Sequential coupling between COPII and COPI vesicle coats in endoplasmic-reticulum to Golgi transport. J Cell Biol. 1995;131:875–893. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. COP II—a membrane coat that forms endoplasmic reticulum-derived vesicles. FEBS Lett. 1995;369:93–96. doi: 10.1016/0014-5793(95)00618-j. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COP II—a membrane coat formed by s proteins that drive vesicle budding from the endoplasmic-reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Birky CW. The partitioning of cytoplasmic organelles at cell division. Int Rev Cytol. 1983;15:49–89. doi: 10.1016/b978-0-12-364376-6.50009-0. [DOI] [PubMed] [Google Scholar]
- Blocker A, Griffiths G, Olivo J-C, Hyman A, Severin F. A role for microtubule dynamics in phagosome movement. J Cell Sci. 1998;111:303–312. doi: 10.1242/jcs.111.3.303. [DOI] [PubMed] [Google Scholar]
- Cassimeris L, Rieder CL, Rupp G, Salmon ED. Stability of microtubule attachment to metaphase kinetochores in PtK1 cells. J Cell Sci. 1990;96:9–15. doi: 10.1242/jcs.96.1.9. [DOI] [PubMed] [Google Scholar]
- Cole NB, Ellenberg J, Song J, DiEuliis D, Lippincott-Schwartz J. Retrograde transport of Golgi-localized proteins to the ER. J Cell Biol. 1998;140:1–15. doi: 10.1083/jcb.140.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption—regeneration of Golgi stacks at peripheral endoplasmic-reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman A, Jones EA, Heasman J. Meiotic maturation in Xenopusoocytes: a link between the cessation of protein secretion and the polarized disappearance of Golgi apparati. J Cell Biol. 1985;101:313–318. doi: 10.1083/jcb.101.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace L, Blobel G. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell. 1980;19:277–287. doi: 10.1016/0092-8674(80)90409-2. [DOI] [PubMed] [Google Scholar]
- Gorman CM, Howard BH. Expression of recombinant plasmids in mammalian cells is enhanced by sodium butyrate. Nucleic Acids Res. 1983;11:7631–7648. doi: 10.1093/nar/11.21.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri HP, Schweizer A. The endoplasmic reticulum-Golgi intermediate compartment. Curr Opin Cell Biol. 1992;4:600–608. doi: 10.1016/0955-0674(92)90078-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshii SO, Yoshida T, Imanakayoshida K, Izutsu K. Distribution of a Ca2+storing site in Ptk2 cells during interphase and mitosis—an immunocytochemical study using an antibody against calreticulin. Eur J Cell Biol. 1995;66:82–93. [PubMed] [Google Scholar]
- Jackman M, Firth M, Pines J. Human cyclins B1 and B2 are localized to strikingly different structures—B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO (Eur Mol Biol Organ) J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano J-I, Soyuki I, Tsutomu O, Suganuma T. A protein-specific monoclonal antibody to rat liver β-1,4 galactosyltransferase and its application to immunohistochemistry. J Histochem Cytochem. 1994;42:363–369. doi: 10.1177/42.3.8308253. [DOI] [PubMed] [Google Scholar]
- Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, Tanigawa G, Rothman JE, Balch WE. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J Cell Biol. 1994;125:51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Cole NB, Marotta A, Conrad PA, Bloom GS. Kinesin is the motor for microtubule-mediated Golgi-to-ER membrane traffic. J Cell Biol. 1995;128:293–306. doi: 10.1083/jcb.128.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Smith C. Insights into secretory and endocytic membrane traffic using green fluorescent protein chimeras. Curr Opin Neurobiol. 1997;7:631–639. doi: 10.1016/s0959-4388(97)80082-7. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq JM, Pryde JG, Berger EG, Warren G. A mitotic form of the Golgi apparatus in HeLa cells. J Cell Biol. 1987;104:865–874. doi: 10.1083/jcb.104.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq JM, Warren G. Fragmentation and partitioning of the Golgi apparatus during mitosis in HeLa cells. EMBO (Eur Mol Biol Organ) J. 1987;6:3239–3246. doi: 10.1002/j.1460-2075.1987.tb02641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- Martinez O, Antony C, Pehau-Arnaudet G, Berger EG, Salamero J, Goud B. GTP-bound forms of rab6 induce the redistribution of Golgi proteins into the endoplasmic reticulum. Proc Natl Acad Sci USA. 1997;94:1828–1833. doi: 10.1073/pnas.94.5.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Koonce MP. Mitosis. Science. 1989;246:622–628. doi: 10.1126/science.2683078. [DOI] [PubMed] [Google Scholar]
- Misteli T, Warren G. COP-coated vesicles are involved in the mitotic fragmentation of Golgi stacks in a cell-free system. J Cell Biol. 1994;125:269–282. doi: 10.1083/jcb.125.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Warren G. Mitotic disassembly of the Golgi apparatus in vivo. J Cell Sci. 1995;108:2715–2727. doi: 10.1242/jcs.108.7.2715. [DOI] [PubMed] [Google Scholar]
- Moskalewski S, Thyberg J. Disorganization and reorganization of the Golgi complex and the lysosomal system in association with mitosis. J Submicrosc Cytol Pathol. 1990;22:159–171. [PubMed] [Google Scholar]
- Mundy DI, Warren G. Mitosis and inhibition of intracellular transport stimulate palmitoylation of a 62-kD protein. J Cell Biol. 1992;116:135–146. doi: 10.1083/jcb.116.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Lowe M, Levine T, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Jackson M, Peterson PA. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell. 1989;58:707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- Pepperkok R, Scheel J, Horstmann H, Hauri HP, Griffiths G, Kreis TE. β-COP is essential for biosynthetic membrane transport from the endoplasmic reticulum to the Golgi complex in vivo. Cell. 1993;74:71–82. doi: 10.1016/0092-8674(93)90295-2. [DOI] [PubMed] [Google Scholar]
- Porter KR, Machado RD. Studies on the endoplasmic reticulum. IV. Its form and distribution during mitosis in cells of onion root tip. J Biophys Biochem Cytol. 1960;7:167–180. doi: 10.1083/jcb.7.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Salmon E. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T, Balch WE. Expression and purification of mammalian Sarl. Methods Enzymol. 1995;257:49–53. doi: 10.1016/s0076-6879(95)57009-8. [DOI] [PubMed] [Google Scholar]
- Scales SJ, Pepperkok R, Kreis TE. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- Schweizer A, Fransen JA, Matter K, Kreis TE, Ginsel L, Hauri HP. Identification of an intermediate compartment involved in protein transport from endoplasmic reticulum to Golgi apparatus. Eur J Cell Biol. 1990;53:185–196. [PubMed] [Google Scholar]
- Seelig HP, Schranz P, Schroter H, Wiemann C, Griffiths G, Renz M. Molecular genetic analyses of a 376-kilodalton Golgi complex membrane protein (giantin) Mol Cell Biol. 1994;14:2564–2576. doi: 10.1128/mcb.14.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol. 1997;137:1211–1228. doi: 10.1083/jcb.137.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G, Georgatos SD. The inner nuclear membrane protein p58 associates in vivo with a p58 kinase and the nuclear lamins. EMBO (Eur Mol Biol Organ) J. 1992;11:4027–4036. doi: 10.1002/j.1460-2075.1992.tb05496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR, Tartakoff AM. The response of the Golgi complex to microtubule alterations: the roles of metabolic energy and membrane traffic in Golgi complex organization. J Cell Biol. 1989;109:2081–2088. doi: 10.1083/jcb.109.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. Membrane partitioning during cell division. Annu Rev Biochem. 1993;62:323–348. doi: 10.1146/annurev.bi.62.070193.001543. [DOI] [PubMed] [Google Scholar]
- Warren G, Wickner W. Organelle Inheritance. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C, Gregory J, Parsons S, Salmon E. Membrane/microtubule tip attachment complexes (TACs) allow the assembly dynamics of plus ends to push and pull membranes into tubulo-vesicular networks in interphase Xenopusegg extracts. J Cell Biol. 1995;130:1161–1169. doi: 10.1083/jcb.130.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EB. The distribution of the chondriosomes to the spermatozoa in scorpions. Proc Natl Acad Sci USA. 1916;2:321–324. doi: 10.1073/pnas.2.6.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Simon PM, Borisy GG. Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J Cell Biol. 1996;135:201–214. doi: 10.1083/jcb.135.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]