Figure 1.

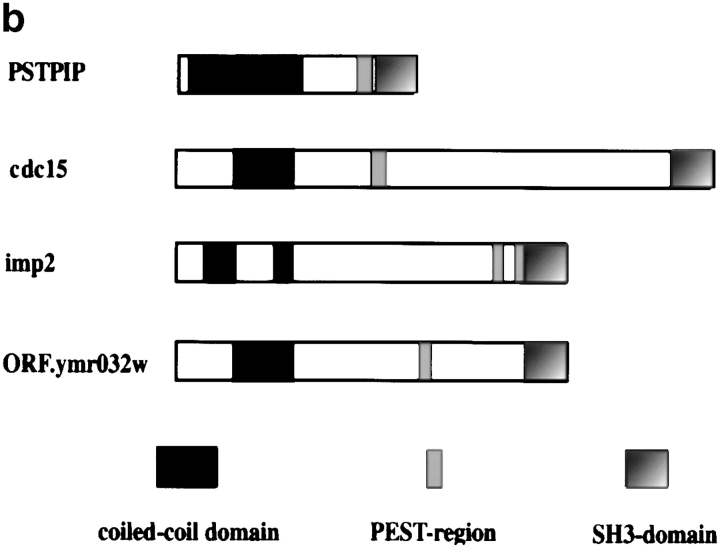
Sequence characteristics of imp2. (a) Amino acid sequence comparison between the predicted products of S. pombe imp2, S. cerevisiae ORF ymr032w, mouse PSTPIP, and S. pombe cdc15. The DNA sequence data imp2 are available from GenBank/EMBL/DDBJ under accession No. Z69379. (b) Schematic representation of domain structure of the proteins encoded by ymr032w, cdc15, imp2, and PSTPIP. Regions showing the highest similarity are the NH2-terminal domains with propensity to form coiled-coil structure and the COOH-terminal SH3 domains. All four proteins have high scoring PEST sequences, but their localization is not conserved. (c) Map of the genomic imp2 locus and the imp2 deletion construct. The internal HindIII fragment of the imp2 gene was replaced by the ura4 + gene carried on a HindIII fragment. Small arrows, oligonucleotides 1–3 with their direction, that were used to verify the null mutation.