Figure 1.
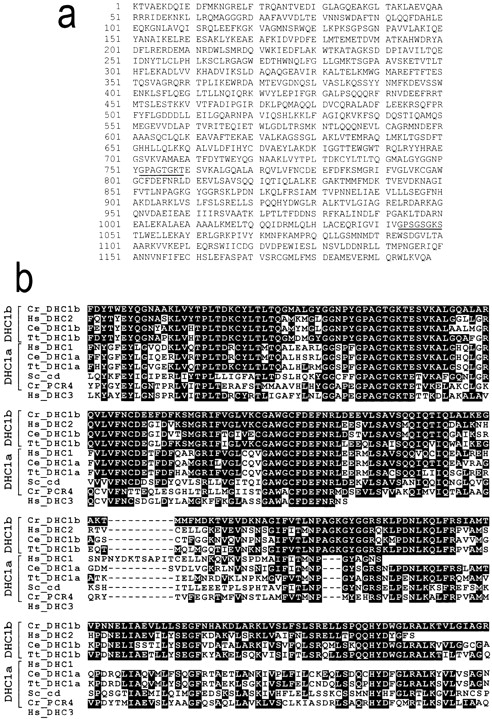
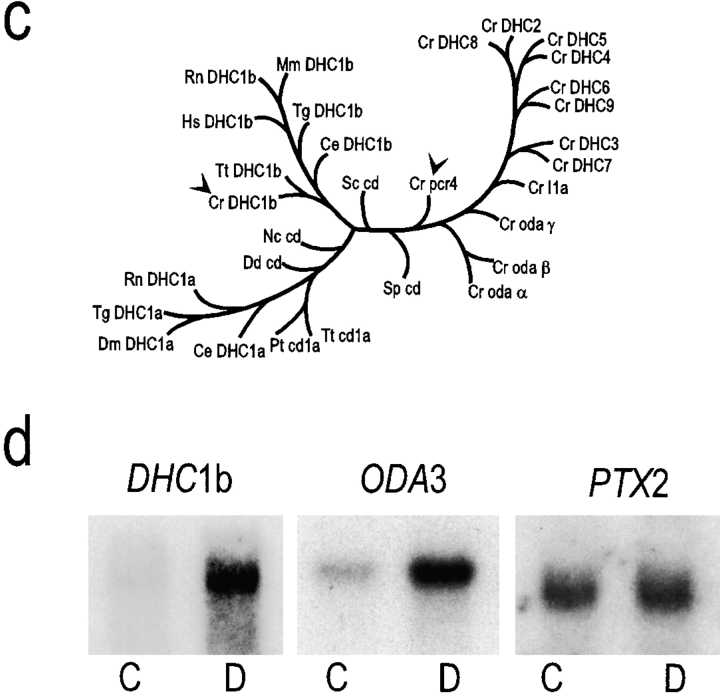
The Chlamydomonas reinhardtii DHC1b gene groups with DHC1b sequences from other organisms and is induced by deflagellation. (a) Partial sequence of the C. reinhardtii gene encoding DHC1b. P-loops 1 and 2 are underlined. Sequence data are available from GenBank/EMBL/DDBJ under accession number AF096277. (b) Alignment of the Chlamydomonas DHC1b and pcr4 peptides with other cytoplasmic dynein heavy chains. A representative selection of DHC sequences from GenBank were aligned with CLUSTAL W and the residues identical to Chlamydomonas DHC1b were shaded in black. Species names are abbreviated as described in c. A longer sequence for pcr4 is available under accession number AF106079. (c) Phylogenetic tree showing the relationship of the Chlamydomonas DHC1b and pcr4 sequences (arrowheads) to other DHC sequences. The predicted peptide sequences of the Chlamydomonas DHC1b cDNA (starting just upstream of P-loop 1 at the sequence CYLTLT and ending with the sequence FVTLNP) and pcr4 cDNA were aligned with a subset of DHC sequences in GenBank using CLUSTAL W, and a phylogenetic tree drawn with PHYLIP using the UPGMA method. Ce, C. elegans; Cr, C. reinhardtii; Dd, Dictyostelium discoideum; Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, Mus musculus; Nc, Neurospora crassa; Pt, Paramecium tetraurelia; Rn, Rattus novegicus; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Tg, Tripneustes gratilla; Tt, Tetrahymena thermophila. (d) The Chlamydomonas DHC1b gene is induced by deflagellation. mRNA was isolated from control, nondeflagellated cells (C), and from cells 30 min after deflagellation (D). The mRNA was analyzed by Northern blot using probes for the DHC1b, ODA3, and PTX2 genes. The latter two genes served as standards; transcription of ODA3 is induced by deflagellation (Koutoulis et al., 1997), whereas that of PTX2, a gene involved in phototaxis, is not (Pazour, G., and G. Witman, unpublished observations).