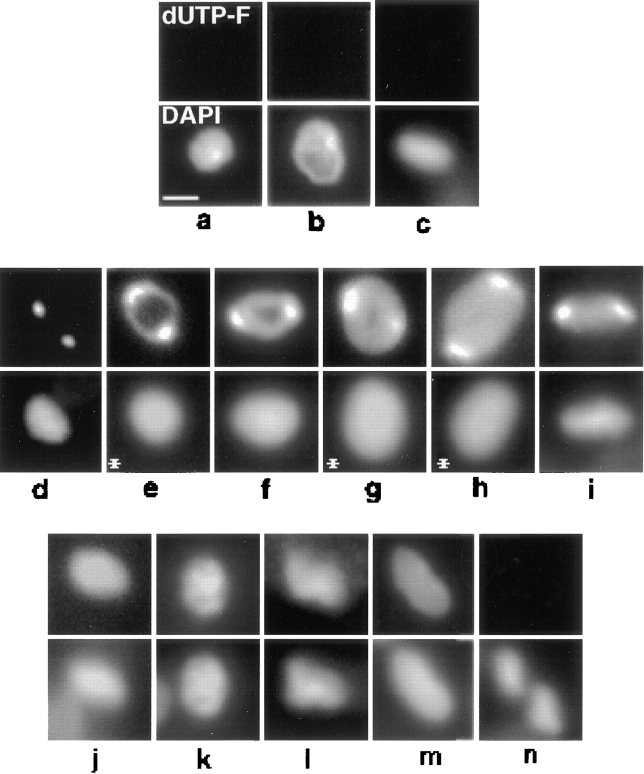
Figure 4.
In situ analysis of kDNA replication in fixed cells from an asynchronous culture. dUTP-F was incorporated into endogenous minicircle gaps by TdT as described in Materials and Methods. Top row, dUTP-F fluorescence of kDNA networks. Bottom row, DAPI fluorescence. (a–c) Pre-replication networks. (d–i) Replicating networks. (j–m) Postreplication networks. (n) Two fully repaired, post scission networks, both in a cell undergoing cytokinesis. The kDNA networks in c and i are positioned vertically. Images were captured with a Photometrics slow-scan CCD camera using IPLab software. Networks in cells treated with proteinase K to reposition the network for a horizontal view are indicated with an asterisk. Treatment with proteinase K was modified from a previous method (Ferguson et al., 1992). Before either immunofluorescence or dUTP-F labeling, fixed cells were treated at room temperature for 15 min with 1 μg/ml Proteinase K in 10 mM Tris-HCl, 1 mM EDTA, 150 mM NaCl, and 0.1% SDS. Slides were then given three 5-min washes in PBS containing 1 mM PMSF, and one 5-min wash in PBS. Without protease treatment, ∼8% of cells contain tipped networks. Treatment with protease increases the frequency of horizontally positioned networks to ∼25%. Bar, 1 μm.