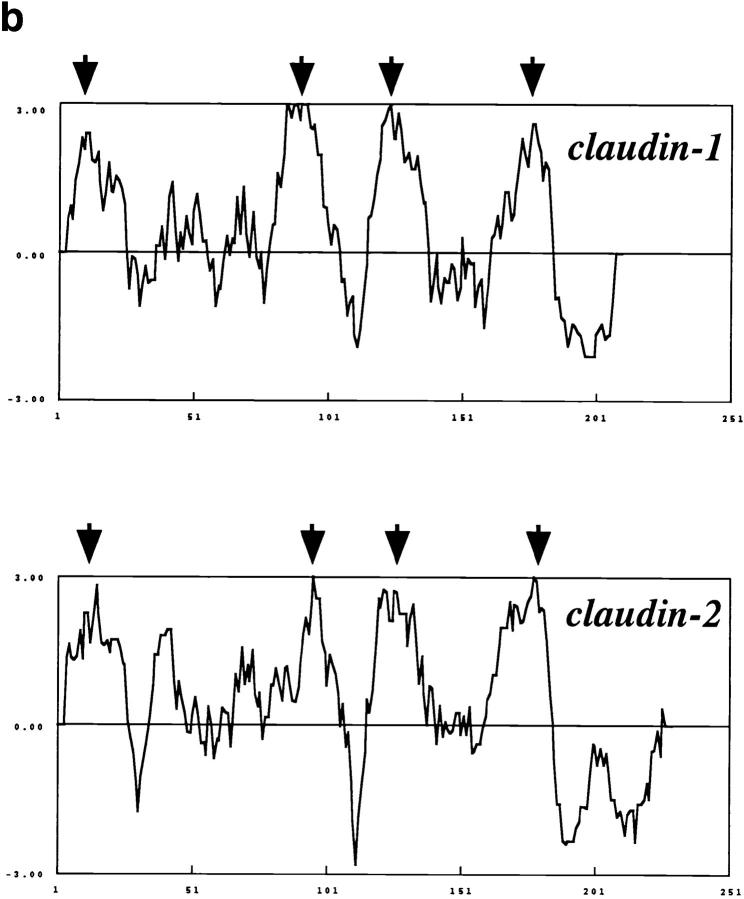
Figure 4.
Structures of mouse claudin-1 and -2. (a) Comparison of amino acid sequences of mouse claudin-1 and -2 by the GENETYX program. Identity and homology are indicated by asterisks and dots, respectively. They showed 38% identity at the amino acid sequence level. (b) Hydrophilicity plots for claudin-1 and -2 prepared using the Kyte and Doolittle algorithm. The plot records the average hydrophilicity along the sequence over a window of eight residues. Hydrophilic and hydrophobic residues are in the lower and upper parts of the frames, respectively. The axes are numbered in amino acid residues. Both claudin-1 and -2 appeared to contain four major hydrophobic, potentially membrane-spanning regions (arrows), although the possibility was not excluded that five transmembrane domains were included in these molecules only from these plots as discussed in the text.