Abstract
The signal recognition particle and its receptor (SR) target nascent secretory proteins to the ER. SR is a heterodimeric ER membrane protein whose subunits, SRα and SRβ, are both members of the GTPase superfamily. Here we characterize a 27-kD protein in Saccharomyces cerevisiae (encoded by SRP102) as a homologue of mammalian SRβ. This notion is supported (a) by Srp102p's sequence similarity to SRβ; (b) by its disposition as an ER membrane protein; (c) by its interaction with Srp101p, the yeast SRα homologue; and (d) by its role in SRP-dependent protein targeting in vivo. The GTP-binding site in Srp102p is surprisingly insensitive to single amino acid substitutions that inactivate other GTPases. Multiple mutations in the GTP-binding site, however, inactivate Srp102p. Loss of activity parallels a loss of affinity between Srp102p and Srp101p, indicating that the interaction between SR subunits is important for function. Deleting the transmembrane domain of Srp102p, the only known membrane anchor in SR, renders SR soluble in the cytosol, which unexpectedly does not significantly impair SR function. This result suggests that SR functions as a regulatory switch that needs to associate with the ER membrane only transiently through interactions with other components.
Keywords: protein targeting, signal recognition particle, Saccharomyces cerevisiae, endoplasmic reticulum, GTPase
Localization of proteins to their correct subcellular compartments is required for eukaryotic cells to maintain the high degree of order required for life. In mammalian cells, protein translocation across the ER membrane is thought to occur primarily in a cotranslational manner. Secretory protein synthesis initiates on cytoplasmic ribosomes, and is completed only after those ribosomes have been targeted to the ER (for reviews see Rapoport et al., 1996; Walter and Johnson, 1994). The signal recognition particle (SRP)1 initially selects cytosolic ribosomes for targeting to the ER if they are synthesizing nascent polypeptides containing NH2-terminal hydrophobic signal sequences. Signal sequences, as they emerge from the ribosome, bind tightly to SRP, causing a transient pause in the translation of the nascent secretory protein. Targeting of this complex to the ER occurs via an interaction between SRP and the SRP receptor (SR), a heterodimeric protein anchored in the ER membrane (Gilmore et al., 1982a ; Gilmore et al., 1982b ; Meyer et al., 1982). This interaction results in localization of the nascent protein to the ER, releases the SRP-induced translational arrest, and allows the nascent chain/ribosome complex to dissociate from SRP/SR. Concomitantly, the ribosome/nascent chain is handed over to the translocon, comprised principally of the Sec61 protein complex (Görlich and Rapoport, 1993; Hanein et al., 1996) that effects translocation of the nascent chain across the lipid bilayer. The translocon forms a protein pore through which the nascent polypeptide crosses the hydrophobic environment of the lipid bilayer (Hanein et al., 1996). Thus, SRP and SR are required for initial targeting of the ribosome/nascent chain to the ER to form the ribosome/translocon junction. The two components therefore couple cytoplasmic translation to the translocation of proteins across the lipid bilayer of the ER membrane.
SRP and SR have been identified in all living cells examined, including eukaryotic cells, bacteria, and archaea (Bult et al., 1996, Walter and Johnson, 1994), indicating that SRP-dependent protein targeting is universally conserved. Identification of homologues of SRP and SR subunits in yeast has allowed study of the role of these components in vivo using primarily reverse genetic approaches (Brown et al., 1995). Studies in yeast have also revealed that in addition to the SRP-dependent cotranslational pathway, an SRP-independent pathway exists that operates in parallel and mediates posttranslational translocation (Hann and Walter, 1991). The signal sequence of the nascent polypeptide determines which route the substrate will take to the ER membrane (Ng et al., 1996).
In eukaryotes, both SRP and SR are multisubunit complexes. SRP is a ribonucleoprotein that consists of six distinct protein subunits and SRP RNA (7SL RNA; Walter and Blobel, 1980; Walter and Blobel, 1982); SR is a heterodimeric ER membrane protein consisting of a 69-kD peripheral membrane protein (SRα) and a 30-kD integral membrane protein (SRβ; Tajima et al., 1986). The best-characterized and phylogenetically most conserved SRP subunit, SRP54, contains the signal sequence–binding activity of SRP, interacts with the SRP RNA, and mediates binding of SRP to SR (Krieg et al., 1986; Kurzchalia et al., 1986, Römisch et al., 1989; Siegel and Walter, 1988; Zopf et al., 1990). In addition, SRP54 contains a GTPase domain (Bernstein et al., 1989; Römisch et al., 1989). Both subunits of SR also contain GTPase domains, and GTP binding to all three GTPases—SRP54, SRα, and SRβ— has been experimentally demonstrated (Connolly and Gilmore, 1986; Connolly and Gilmore, 1989; Miller et al., 1995). Thus, three distinct directly interacting GTPases collaborate in protein targeting, and the elucidation of the mechanisms with which their respective GTPase cycles control the targeting events is of considerable interest.
The GTPase domains of SRP54 and SRα are related, defining their own subfamily in the GTPase superfamily as recently underscored by the elucidation of the crystal structure of their prokaryotic homologues (Freymann et al., 1997; Montoya et al., 1997). According to a current model for the cycle of GTP binding and hydrolysis of SRP54 and SRα, as SRP54 binds to the signal sequence of a nascent secretory protein, it becomes stabilized in a state displaying a low affinity for GTP (Miller et al., 1993; Rapiejko and Gilmore, 1997). Interaction with the ribosome may enhance SRP54's affinity for GTP (Bacher et al., 1996). Targeting of this complex to the ER membrane and interaction of SRP with SR effects a further increase in the affinity of SRP54 for GTP. The resulting change in conformation of SRP54 is thought to accompany release of the signal sequence from SRP, leaving the ribosome/nascent chain complex free to interact with components of the translocon. SRP then remains associated with SR after release of the ribosome/nascent chain complex until hydrolysis of the GTPs bound to SRP54 and SRα results in dissociation of SRP from SR. Hydrolysis occurs by an enzymatically reciprocal interaction, as SRP54 and SRα act as GTPase-activating proteins for each other while they are in the GTP-bound state (Powers and Walter, 1995), possibly reflecting the structural relatedness of the two interacting GTPase domains.
The role of the GTPase domain in SRβ has remained more enigmatic. The GTPase domain of SRβ is structurally distinct from those of SRα and SRP54; it falls into its own subfamily of GTPases with its closest, albeit still relatively distant relative being Sar1p, an ARF-like GTPase involved in ER-to-Golgi trafficking (Miller et al., 1995). We describe here the first functional characterization of yeast SRβ (Srp102p) encoded by the SRP102 gene. We show that Srp102p anchors Srp101p (the yeast SRα homologue) to the ER membrane, consistent with its disposition as an integral ER membrane protein. Unexpectedly, however, a mutant form of Srp102p lacking the transmembrane domain is functional in yeast, indicating that Srp102p is not required solely to tether Srp101p to the membrane. In contrast, mutations that sufficiently disrupt the GTP-binding site of Srp102p inactivate SR. Most of these mutants also disrupt the interaction between Srp101p and Srp102p, suggesting that an interaction of Srp102p with Srp101p may be required to activate Srp101p as a prerequisite for a productive interaction with SRP during protein targeting.
Materials and Methods
Strains, Antibodies, Materials, and General Methods
Yeast strains are listed in Table I. Genetic techniques are performed as described previously, except where noted (Sherman and Lawrence, 1974). Yeast transformation was performed by the lithium acetate procedure (Ito et al., 1983). Yeast DNA for Southern analysis was prepared as described (Davis et al., 1980). Recombinant DNA techniques and Southern blots were performed as described (Sambrook et al., 1989). Western blots were visualized using enhanced chemiluminescence (Amersham Corp., Arlington Heights, IL) as described by the manufacturer. Oligonucleotides were synthesized on a Cyclone Plus™ DNA synthesizer (Millipore, Novato, CA) and purified according to the manufacturer's instructions. Both high and low-copy yeast shuttle vectors used are as described (Christianson et al., 1992, Sikorski and Heiter, 1989) unless otherwise indicated. Chemicals are from Sigma Chemical Co. (St. Louis, MO) unless otherwise noted. The plasmid library used for cloning SRP102 was obtained from the American Type Culture Collection (catalog no. 37415; Rockville, MD). Anti-Kar2p serum was prepared in our laboratory using Kar2p overexpressed in Escherichia coli from a clone kindly provided by Joe Vogel and Mark Rose (Rose et al., 1989). 0.5 μl of anti-Kar2p serum was used per OD600 in nonnative immunoprecipitations. Fluorochrome-coupled anti-rabbit IgG and anti-mouse IgG secondary antibodies are from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). Anti-Srp101p is used as described in Ogg et al. (1992). Monoclonal 12CA5 (anti-HA) and 9E10 (anti-myc) were purchased from Berkeley Antibody Company (Richmond, CA), and were used at a dilution of 1:10,000 on Western blots. DNA sequencing was performed using an ABS A220 fluorescent DNA sequencer (Perkin-Elmer Corp., Norwalk, CT) and cycle sequencing.
Table I.
Yeast Strains and Plasmids Used in This Study
| Strain | Genotype | Reference | ||||
|---|---|---|---|---|---|---|
| W303 | ade2-1/ade2-1 trp1-1/trp1-1 leu2-3, 112/leu2-3, 112 his3-11/his3-11 ura3-1/ura3-1 | Deshaies and Schekman, 1990 | ||||
| can1-100/can1-100 MATa/MATα | ||||||
| TR1 | trp1/trp1 lys2/lys2 his3/his3 ura3/ura3 ade2/ade2 MATa/MATα | Parker et al., 1988 | ||||
| TR2 | trp1 lys2 his3 ura3 ade2 MATa | Parker et al., 1988 | ||||
| TR3 | trp1 lys2 his3 ura3 ade2 MATα | Parker et al., 1988 | ||||
| SOY133 | W303 MATa/MATα SRP102/srp102::HIS3 | This study | ||||
| SOY162 | W303 MATα srp102::HIS3 | This study | ||||
| SOY195 | W303 MATα srp101::ADE2, srp102::HIS3 | This study | ||||
| SOY246 | SOY162 [pSO462] | This study | ||||
| SOY223 | SOY162 [pSO459] | This study | ||||
| SOY201 | SOY162 [pSO454] | This study | ||||
| SOY264 | SOY162 [pSO462] [pSO211] | This study | ||||
| SOY268 | SOY162 [pSO462] [pSO400] | This study | ||||
| SOY329 | TR1 SRP102/srp102::HIS3 | This study | ||||
| SOY335 | TR3 srp102::HIS3 [pSO459] | This study | ||||
| WBY338 | WBY618 [pWB209] | This study | ||||
| WBY618 | TR2 srp102::HIS3 | This study | ||||
| WBY632 | TR3 srp102::HIS3 [pSO452] | This study | ||||
| WBY752 | TR3 srp101::ADE2 srp102::HIS3 [pSO431] [pWB209] | This study | ||||
| WBY823 | TR2 srp101::ADE2 srp102::HIS3 [pWB209] | This study | ||||
| Plasmids | Markers | Backbone | Reference | |||
| pSO452 | URA3, SRP102, CEN6/ARSH4 | pRS316 | Sikorski and Heiter, 1989 | |||
| pSO454 | TRP1, SRP102, CEN6/ARSH4 | pRS314 | Sikorski and Heiter, 1989 | |||
| pSO457 | TRP1, SRP102-myc, REP3 | YEplac112 | Geitz and Sugino, 1988 | |||
| pSO431 | URA3, SRP101-HA, CEN6/ARSH4 | pRS316 | Sikorski and Heiter, 1989 | |||
| pSO459 | TRP1, SRP102-HA, CEN6/ARSH4 | pRS314 | Sikorski and Heiter, 1989 | |||
| pSO462 | TRP1, srp102(K51I)-HA, CEN6/ARSH4 | pRS314 | Sikorski and Heiter, 1989 | |||
| pSO211 | URA3, SRP101, CEN6/ARSH4 | pRS316 | Sikorski and Heiter, 1989 | |||
| pSO400 | URA3, pGAL-SRP101, CEN6/ARSH4 | pRS316 | Sikorski and Heiter, 1989 | |||
| pWB670 | TRP1, srp102-ΔTMD-HA, CEN6/ARSH4 | pRS314 | Sikorski and Heiter, 1989 | |||
| pWB209 | TRP1, srp102-ΔTMD, CEN6/ARSH4 | pRS314 | Sikorski and Heiter, 1989 |
Cloning of SRP102
Initial attempts to clone SRP102 using PCR from genomic DNA, and the known sequence to design oligonucleotides resulted in the clones having multiple sequence errors, presumably from inaccurate PCR. Therefore, a genomic clone of SRP102 was isolated by screening a plasmid library prepared from Saccharomyces cerevisiae strain GRF88 in the vector YCp50 using one of the PCR-obtained clones as a probe to screen the library (Rose et al., 1987). One of the plasmids obtained in the initial PCR attempt to clone SRP102 was random-primed using the Ready To Go™ DNA labeling kit according to the manufacturer's instructions (Pharmacia Biotech, Inc., Piscataway, NJ). Three plasmids were isolated from the library screen (designated pSO451a, b, and c) that contained identical inserts, and the 2966-bp EcoRI fragment predicted to contain SRP102 was subcloned into the EcoRI sites of pRS314, pRS316, and pRS426 yielding plasmids pSO454, pSO452, and pSO453, respectively. Both strands of the insert in pSO452 were sequenced from nucleotides 175– 821, and this sequence agreed with the published genomic sequence. Confirmation that these plasmids contained a functional open reading frame was obtained by complementation of the SRP102 gene disruption.
Gene Disruption
We generated a null allele of SRP102 in a two-step fashion. First we used PCR to generate DNA fragments corresponding to the 5′ and 3′ flanking regions of SRP102, and subcloned these into pBluescript II SK (+) (Stratagene, La Jolla, CA), engineering a SmaI site at the junction between the flanking sequences. The HIS3 gene was then inserted into the SmaI site, generating pSO446, now carrying the HIS3 gene between the 5′ and 3′ flanking regions of SRP102. A linear fragment carrying the SRP102 disruption allele was generated and used to transform the diploid yeast strain W303, giving rise to strain SOY133. Transformants were induced to sporulate. Genomic DNA prepared from the heterozygous diploid (SOY133), wild-type parent (W303), and representative spores from a single tetrad were used as template in PCR. When DNA from the wild-type parent and the His− spores was used as template in the PCR, the expected ∼1.42-kb fragment was seen. Use of the DNA isolated from the His+ spores gave rise to a single expected ∼2.65-kb fragment. Only when DNA prepared from the heterozygous diploid was used as template in the reaction were both the 1.42-kb and the 2.65-kb fragments seen.
Plasmid and Strain Construction
To construct a plasmid containing SRP102 under control of the GAL1/10 promoter, we used PCR to produce a fragment corresponding to the SRP102 coding sequences, and subcloned this fragment into pTS210 (plasmid kindly provided by Tim Stearns, Stanford University, Palo Alto, CA), generating pSO460. This plasmid was transformed into SOY133, and the resulting Ura+ prototrophs were induced to sporulate. Asci were dissected, and spores were allowed to germinate at room temperature on YEP plates containing 2% galactose and 2% sucrose as carbon sources. Colonies that were both Ura+ and His+ were chosen for further study. The fact that His+, Ura+ colonies arose that grew with wild-type rates supports our assignment of the initiation codon, as Srp102p expressed from pSO460 starts at this codon. Additionally, these His+, Ura+ colonies were incapable of growing for more than a few generations on plates containing 2% glucose as the sole carbon source, indicating that we had indeed placed SRP102 under conditional control.
To generate the strains we used in our fractionation and immunofluorescence studies, we placed plasmids containing Srp102p (pSO454, no epitope tag), or Srp102p-HA (pSO459), or Srp102p-myc (pSO457) into SOY133 (W303 background) or SOY329 (TR1 background), and induced the diploid transformants to sporulate. Spores giving rise to colonies that were both His+ (SRP102::HIS3) and Trp+ (containing a plasmid) and that grew at wild-type rates were chosen for further study, and these strains were designated as indicated in Table I. We constructed pSO457 in the following manner: a PCR product containing the SRP102-coding sequence was ligated into pCS118 (plasmid kindly provided by Caroline Shamu, Harvard University, Boston, MA). pCS118 contains a 400-bp insert harboring the myc tag, stop codons in all three reading frames, and an actin transcriptional terminator in YEplac112 (Geitz and Sugino, 1988). Thus, pSO457 contains the SRP102 coding sequence fused at the COOH terminus to the myc tag followed by an actin terminator, all in the YEplac112 (TRP1, 2μ) backbone. A similar strategy was used to construct pSO459 using pSF19 (plasmid kindly provided by Peter Sorger, Massachusetts Institute of Technology, Boston, MA) as the recipient vector. This plasmid contains SRP102-HA in the pRS314 backbone (TRP1, CEN/ARS).
To construct the ΔTMD strain, 66 bp of SRP102 (coding for amino acids 3–24 of Srp102p) were deleted from pSO454 by recombinant PCR (Higuchi et al., 1988) using Vent polymerase (New England Biolabs, Beverly, MA). Both strands of the truncated SRP102 gene of this plasmid were sequenced to show that only the desired deletion had been introduced. The diploid heterozygous strain SOY329 was transformed to Trp+ prototrophy with pWB209, and a transformant was induced to undergo meiosis. Tetrads were dissected and allowed to germinate on minimal plates lacking tryptophan. All His+ spores (containing the srp102::HIS3 deletion allele) that were also Trp+ (containing pWB209, the ΔTMD allele) gave rise to colonies of similar size as the His− colonies (containing the wild-type SRP102 allele).
To replace the transmembrane domain (TMD) of Srp102p with an HA epitope tag, recombinant PCR was used in the same manner as described for the construction of pWB209. The resulting plasmid, pWB670, encodes a mutated Srp102p protein with an NH2-terminal HA tag instead of the TMD domain. We replaced pSO452 in strain WBY632 with pWB670 using the plasmid-shuffle technique, generating strain WBY670.
Preparation of Antibodies Specific to Srp102p
Antiserum was raised against a fusion protein expressed in E. coli, containing glutathione-S-transferase from S. japonicum as the NH2-terminal domain linked to the soluble domain of Srp102p. To generate this fusion protein, a DNA fragment coding all but the NH2-terminal 24 amino acids of Srp102p was amplified by PCR using Vent polymerase (New England Biolabs, Beverly, MA), generating the fusion protein plasmid, pSO488.
The GST-Srp102p fusion protein, containing amino acids 25–244 of Srp102p, was soluble when it was overexpressed in E. coli at 24°C. Cells were lysed by sonication, and the fusion protein was purified using glutathione-agarose (Sigma Chemical Co.) as described (Smith and Johnson, 1988). Polyclonal rabbit antiserum against the fusion protein was prepared by Berkeley Antibody Company (Richmond, CA). Antisera were used without further purification.
Site-directed Mutagenesis
Site-directed mutagenesis was performed by the method of Kunkel (Kunkel, 1985; Kunkel et al., 1987). Single-stranded uracil-rich DNA used for template was prepared from the phagemid pSO459 (containing SRP102-HA). Plasmids that were recovered from a mutagenesis reaction, and that were subsequently confirmed to have incorporated the desired mutation, are designated with the wild-type amino acid in single letter code, position of the amino acid changed, and the name of the new amino acid in single letter code (e.g., S49A signifies that a serine at position 49 has been changed to an alanine). Every mutation was confirmed by sequencing the recovered plasmid. The following is a compilation of each of the mutations obtained, and the corresponding oligonucleotide that was used to produce that mutation (wild-type sequences are in normal type and the codon changed is in bold; nucleotide(s) that are noncomplementary to the rescued strand of pSO459 are shown underlined): pSRP102-S49A (oSRP102-25) 5′-CGT TTT TCC AGC ATT TTG AGG-3′; pSRP102-K51I (oSRP102-16) 5′-G CAA GCT CGT AAT TCC AGA ATT TTG-3′; pSRP102-T52N (oSRP102-17) 5′-CGT AAG CAA GCT ATT TTT TCC AGA-3′; pSRP102-T66A (oSRP102-22) 5′-GA AAC AAC AGC TGG TCT TAC-3′; pSRP102-G90L (oSRP102-18) 5′-CTT GAC ATG CAA TGG GAA GTC-3′; pSRP102-H91L (oSRP102-20) 5′-CAA CTT GAC CAA GCC TGG GAA GT-3′; pSRP102-N154I (oSRP102-19) 5′-C GCT TTT AAT GCA TGC AAT TAA G-3′; pSRP102-E157Q (oSRP102-15) 5′-GT GAA CAA TTG GCT TTT ATT GC-3′; pSRP102-S220A (oSRP102-23) 5′-GC AAC TAC AGC TGC TTC C-3′; pSRP102-K51A, T52A (oSRP102-21) 5′-CGT AAG CAA GCT AGCAGC TCC AGA ATT TTG-3′; pSRP102-N154A, E157A (oSRP102-24) 5′-GC AGT GAA CAA AGC GCT TTT AGC GCA TGC AAT TAA G-3′; pSRP102-ΔS49 (oSRP102-26) 5′-CT CGT TTT TCC AGC AGCAGC ACC TGC AAT G-3′; pSRP102-K51I, H91L (oSRP102-16 + oSRP102-20) see above; pSRP102-K51I, N154I (oSRP102-16 + oSRP102-19) see above; pSRP102-K51A,T52A, H91L (oSRP102-21 + oSRP102-20) see above; pSRP102-K51A, T52A, E157Q (oSRP102-21 + oSRP102-15) see above; pSRP102-H91L, N154I (oSRP102-20 + oSRP102-19) see above; pSRP102-H91L, N154A/E157A (oSRP102-20 + oSRP102-24) see above; pSRP102-H91L, E157Q (oSRP102-20 + oSRP102-15) see above.
Cell Fractionation
Cell fractionation by differential centrifugation was performed as described in Ogg et al. (1992) with the following modifications. Zymolyase-100T (ICN Biomedicals, Costa Mesa, CA) instead of lyticase was used at 5 μg/OD600 to convert cells into spheroplasts. Only PMSF and leupeptin were included as protease inhibitors. Whole-cell lysate cleared of unbroken cells and nuclei (low-speed supernatant, see below) was used as the starting material for separation of the ER membrane from cytosol using isopycnic centrifugation. 2.5 OD600 cell equivalents of the low-speed supernatant (50 μl of 50 OD600/ml) was diluted with 200 μl 2.5 M sucrose in flotation buffer (FB; 10 mM Hepes-KOH, pH 7.5, 150 mM KOAc, pH 7.5, 5 mM Mg(OAc)2, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.5 μg/ml leupeptin) to 10 OD600/ml. This dilution served to raise the density of the low-speed supernatant so that it could be placed underneath a discontinuous sucrose gradient. Gradients were formed in TLS-55 polycarbonate tubes (Beckman, Palo Alto, CA). Each gradient consisted of the following from top to bottom: 0.6 ml of 0.8 M sucrose in FB; 1.6 ml of 1.8 M sucrose in FB; 0.2 ml of the load described above, equal to 2.0 OD600 U. Gradients were centrifuged at 55,000 rpm in the TLS-55 rotor in a TL100 tabletop ultracentrifuge for 4 h. Each gradient was fractionated into 10 240-μl fractions, and proteins in each fraction was precipitated with ice-cold TCA added to a final concentration of 10% before analysis by Western blot.
Immunofluorescence
Immunofluorescence was performed essentially as described (Pringle et al., 1989). Anti-HA, and anti-myc antibodies were used at dilutions of 1:200 to 1:2,000. Anti-Kar2p antibodies were used at a dilution of 1:10,000. Bound primary antibodies were decorated with either FITC-conjugated donkey anti–mouse IgG or TRITC-conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) diluted 1:50. Slides were mounted in 90% glycerol containing 1 mg/ml p-phenylenediamine, pH 9.0, and containing 1 mg/ml 4′, 6-diamino-2-phenylindole (DAPI).
Immunoprecipitations
Denaturing immunoprecipitations were performed as described (Ogg et al., 1992). Nondenaturing immunoprecipitations were performed from detergent-solubilized membranes in the following manner: cells were grown to 0.5 OD600/ml in synthetic complete medium lacking amino acids used for maintenaning a plasmid containing SRP102-HA. Typically, 30 total OD600 were harvested at a time. After pelleting in a clinical centrifuge and washing with 50 mM NaCl, 20 mM NaN3, cells were resuspended in buffer A (Tris-SO4, pH 9.4, 10 mM DTT) at 10 OD600/ml, and were incubated at room temperature for 20 min. Conversion of the cells to spheroplasts was then achieved by harvesting the cells in a clinical centrifuge and resuspending them in spheroplast media (50 mM Tris-Cl, pH 7.5, 1.2 M Sorbitol, 5 mM MgCl2, 5 mM DTT, 10 mM NaN3) at 20 OD600/ml. We used 5 μg/OD600 Zymolyase 100-T (ICN Biomedicals, Costa Mesa, CA) to digest the cell walls during a 30-min incubation at 30°C. Efficiency of spheroplasting was monitored by phase contrast microscopy. Spheroplasts were recovered by centrifugation through a cushion (twice the volume of the spheroplasting cells) of spheroplast media containing 1.9 M sorbitol but no DTT at 7,800 g for 5 min (7,000 rpm in a Beckman JS-13 rotor). All subsequent steps were performed at 4°C. Spheroplasts were resuspended in lysis buffer (LB; 200 mM Sorbitol, 25 mM NaPi, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM PMSF, 0.5 μg/ml of leupeptin, 1 mM EDTA) at 100 OD600/ml. Cells were lysed by adding 3/4 vol of Zirconium beads (BioSpec Products, Bartlesville, OK) and vortexing gently (setting 3 on a scale of 1–10) for four rounds of lysis. Each round consisted of 15 s of vortexing followed by a 15-s cooling period in an ice-water bath. Lysate was removed from the beads, which were washed once with ice-cold LB. The lysate and bead wash were combined, and the unbroken cells and nuclei were removed by centrifugation at 370 g (1,200 rpm in a JS-13) for 4 min. This step was critical; longer centrifugation resulted in loss of most of the ER as judged by the pelleting of an ER marker, Srp101p. The low-speed supernatant containing crude membranes was transferred to TLA100.1 polycarbonate tubes, and membranes were pelleted by centrifugation at 43,630 g for 12 min (35,000 rpm in the Beckman TLA100.1 rotor in a Beckman TL100 tabletop ultracentrifuge). The membrane pellet was washed (30 OD600/ml) with LB. This wash was achieved by first resuspending the pellet in a minimal volume of LB (20–40 μl), followed by dilution to the appropriate volume with LB. Again, the membranes were pelleted by centrifugation in the TL100 as above. The final membrane pellet was resuspended in LB + 10% glycerol (GLB) to 50 OD600/ml using the same technique as described above to ensure even membrane resuspension. Membranes were solubilized by adding 1/10th vol of 10% Triton X-100 (Calbiochem, La Jolla, CA) and incubation at 4°C for 20 min. After incubation, the detergent-solubilized membranes were cleared by centrifugation at 100,000 g in the TL 100 centrifuge (60,000 rpm in the TLA100.1) for 25 min. The resulting lysate was frozen in liquid nitrogen and stored at −80°C.
Immunoprecipitations were performed by diluting 2 OD600 U (40 μl) of the above lysate into 560 μl of GLBT (GLB containing 1% Triton X-100). Nonspecific binding of proteins in the lysate to Sepharose was removed by adding 30 μl of a 50% slurry of Sepharose CL-4B (Pharmacia Biotech Sverige, Uppsala, Sweden) and rotating for 30 min at 4°C. Sepharose beads were removed by centrifuging in an Eppendorf microfuge and transferring the lysate to a new tube. Antibody–sepharose bead complexes (10 μl; corresponding to 3 μl of the 12CA5 ascites fluid) were added, and the tube was mixed by rotating at 4°C for 15 min. We found that with the 12CA5 antibody, longer incubation times of the antibody– bead complex resulted in lower recovery of Srp101p in the immunoprecipitation. The 12CA5 antibody used in the native immunoprecipitations was covalently attached to a Sepharose matrix by using the Affinica antibody coupling kit (Schleicher & Schuell, Inc., Keene, NH) and the manufacturer's instructions. After extensive washing, the antibody bead complex was stored as a 50% slurry in PBS containing 10 mM NaN3.
Results
Disruption and Cloning of SRP102
Database searches revealed an open reading frame (YKL154w) encoding a putative GTPase of unknown function on S. cerevisiae chromosome XI that has significant structural similarity to mammalian SRβ (Miller et al., 1995). Data presented below confirm that this gene indeed encodes the yeast homolog of mammalian SRβ and plays an essential role in the SRP-dependent protein-targeting pathway to the ER membrane. Henceforth, we refer to this gene encoding the β-subunit of the SRP receptor as SRP102, consistent with the designation of the gene encoding the α-subunit of the SRP receptor as SRP101 (Ogg et al., 1992). By convention, we shall refer to the corresponding gene products as Srp102p and Srp101p, respectively.
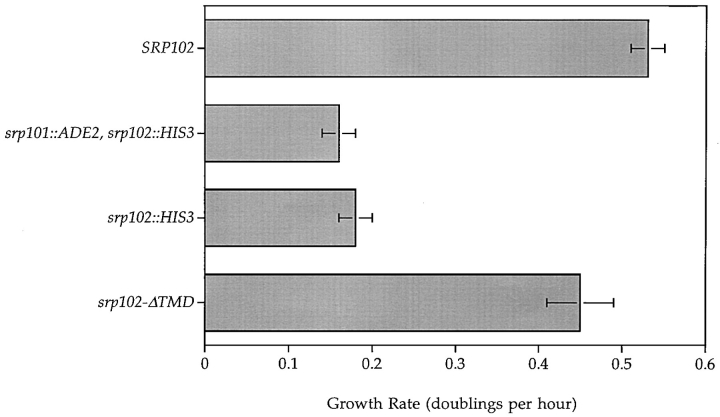
As the first step towards a molecular genetic characterization of SRP102, we examined the phenotype of a gene deletion. To this end we replaced the central portion of one copy of SRP102 in diploid cells with the selectable marker HIS3 (see Methods). Tetrad analysis after sporulation of this strain resulted in four viable spores, two of which grew at wild-type rates while the other two grew with a much reduced rate (Fig. 1, SRP102 vs. srp102:: HIS3). In each of seven tetrads analyzed, the growth defect, the His+ phenotype, and the srp102::HIS3 gene disruption (as monitored by PCR analysis; see Methods) always segregated 2:2 from the SRP102 wild-type allele. Thus, we conclude that the loss of SRP102 was directly responsible for the growth defect.
Figure 1.
Cell growth rates of srp102 mutant strains. Growth rates in YEPD medium at 30°C were determined for wild-type (SRP102, WBY618 bearing pSO452) cells, cells in which the chromosomal SRP102 gene is deleted alone (srp102::HIS3, WBY618) or in combination with the SRP101 gene (srp101::ADE2, srp102::HIS3, SOY195), or in which the chromosomal deletion is covered by srp102-ΔTMD on a plasmid (srp102-ΔTMD, WBY618 bearing pWB209).
We isolated a plasmid carrying the functional SRP102 gene from a yeast genomic DNA library (see Materials and Methods). An EcoRI fragment containing the entire open reading frame and flanking sequences was subcloned, generating plasmid pSO452 (URA3). To show that the information contained in this sequence was sufficient to complement the srp102::HIS3 gene disruption, plasmid pSO452 was transformed into diploid SRP102/srp102:: HIS3 cells. After sporulation, asci were dissected, and the colonies resulting from germination of each of the four spores were analyzed. In all tetrads analyzed (12), each spore that contained the srp102::HIS3 gene disruption (as detected by the His+ phenotype) and that grew at wild-type rates also contained plasmid pSO452 (as detected by the Ura+ phenotype). This result confirms that pSO452 is required for srp102::HIS3 cells to grow at wild-type rates, and thus contains all the information necessary for functional expression of the SRP102.
The slow growth phenotype of cells bearing the srp102:: HIS3 gene disruption closely resembles that of cells in which SRP101 or any of the seven known genes encoding SRP subunits are disrupted. Furthermore, srp102::HIS3 mutant cells—like yeast cells depleted of other SRP components—are rho−, and thus require a fermentable carbon source. Finally, yeast cells in which both SRP101 and SRP102 (Fig. 1) or SRP54 and SRP102 (not shown) are disrupted have a growth defect indistinguishable from cells bearing any single gene disruption, consistent with the notion that these genes operate in the same pathway. Taken together, with the significant sequence similarities between Srp102p and mammalian SRβ, these results strongly suggest that Srp102p is an essential component of the SRP-dependent targeting pathway in S. cerevisiae.
Srp102p is Important for Translocation of Proteins into the ER
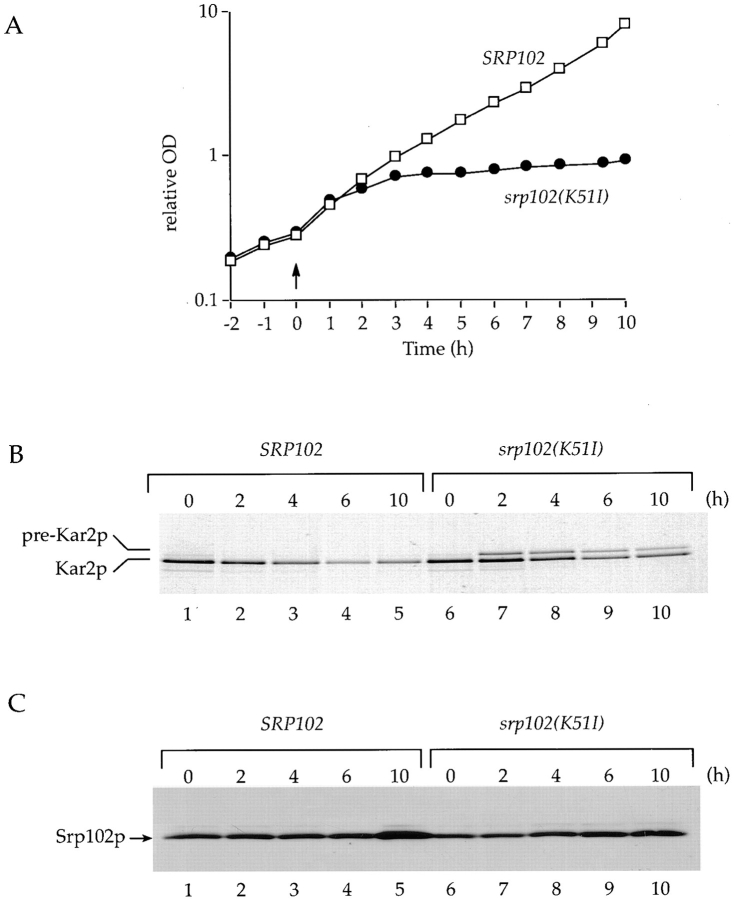
To show directly that Srp102p plays a role in the SRP- dependent protein translocation pathway, we asked if protein translocation defects could be detected upon Srp102p inactivation. To this end, we used a temperature-sensitive allele of SRP102, srp102(K51I), that was isolated by site- directed mutagenesis (see below). Haploid cells containing the chromosomal srp102::HIS3 gene disruption and bearing a plasmid that encodes the srp102(K51I) mutant allele were shifted from the permissive (24°C) to the restrictive temperature (37°C). The growth curve indicates that within 2 h after the temperature shift, the strain carrying the srp102(K51I) allele decreases its growth rate by about sixfold (Fig. 2 A). This difference in doubling time (2 vs. 12 h, respectively) resembles the difference between the wild-type and the isogenic srp102::HIS3 deletion strain (Fig. 1), indicating that Srp102p is inactivated upon temperature shift (note that the growth differences in Fig. 1 are less pronounced, however, because cells were grown in rich media.)
Figure 2.
Srp102p is important for protein translocation. (A) Growth curves. Growth of a wild-type strain (SOY162, srp102:: HIS3 covered by pSO459; open squares) compared with a strain containing the srp102(K51I) ts allele of SRP102 (SOY162, srp102::HIS3 covered by pSO462; closed circles). At time zero (arrow) a culture in midlogarithmic growth of each of the strains growing in synthetic media lacking histidine and tryptophan (to select both for the deletion and the plasmid expressing either the wild-type Srp102p or Srp102[K51I]p) was shifted from 24°C to 37°C. Cell growth was monitored by change in the OD600 of the culture. (B) Kar2p immunoprecipitation. Cells grown as in A were labeled with [35S]methionine for 7 min, and were harvested at the times indicated after a shift from 24°C to 37°C and processed for immunoprecipitation. Lysates at each of the time points indicated were immunoprecipitated with anti-Kar2p and subjected to SDS-PAGE followed by fluorography. Position of both mature and precursor forms of Kar2p are indicated. (C) Western blot. 0.5 OD600 cell equivalents of the lysate prepared in B were subjected to SDS-PAGE and subsequent transfer to nitrocellulose. Srp102p-HA or Srp102(K51I)p-HA was detected with the anti-HA monoclonal antibody, and was visualized by enhanced chemiluminescence.
To monitor the effect of Srp102p inactivation on protein translocation, we pulse-labeled srp102(K51I) cells with [35S]methionine at different time points after shifting to the restrictive temperature. Lysates from labeled cells were analyzed by immunoprecipitation with antibodies raised against proteins that are known to be translocated across the ER membrane. Using antibodies specific for Kar2p, an ER lumenal protein, accumulation of the untranslocated precursor form preKar2p was detected (Fig. 2 B, lanes 7–10). The slower mobility of preKar2p in SDS-PAGE results from the presence of its signal sequence, which is normally cleaved upon entry into the ER. Significant amounts of pre-Kar2p are apparent at the earliest time point taken after placing the cells at the restrictive temperature (2-h time point in Fig. 2 B, lane 7). In similar experiments, we have detected a comparable fraction of preKar2p as early as 30 min after the temperature shift. As expected, no preKar2p was detected in the control strain carrying the wild-type allele of SRP102 (Fig. 2 B, lanes 1–5). Note that at later time points after the temperature shift, the translocation defect of Kar2p is diminished (Fig. 2 B, lanes 8–10). This adaptation of cells in response to the loss of the SRP-dependent targeting pathway was previously observed after shut-off of the synthesis of SRP components and Srp101p (Ogg et al., 1992). Adaptation is a physiological change in the cells that either improves the efficiency of the alternative targeting pathway or helps degrade cytosolic precursor molecules more efficiently.
The severity of translocation defects varies widely for different proteins in the strains from which Srp101p or SRP components are depleted (Hann and Walter, 1991; Ogg et al., 1992). We obtained similar results when radiolabeled extracts of temperature-shifted cells bearing the srp102(K51I) allele were analyzed for translocation defects of two additional proteins, dipeptidyl aminopeptidase B (DPAP-B), a vacuolar membrane protein, and carboxypeptidase Y (CPY), a soluble vacuolar protein (not shown). The membrane integration of DPAP-B, like Kar2p, was significantly affected as Srp102p was rendered nonfunctional upon temperature shift, whereas the translocation of CPY was completely unaffected. These results closely mimic those obtained upon inactivation of other gene products that function in the SRP-dependent protein targeting pathway and lend further support to the notion that Srp102p functions as an essential subunit of the SRP receptor in the same pathway.
We monitored the fate of mutant Srp102p by Western blotting to determine if the loss of Srp102p activity is due to the degradation of the protein at the restrictive temperature. To this end, we took advantage of an HA-epitope tag (see below) that was added to the COOH-terminal end of Srp102p during construction of the mutant allele. Western blot analysis of lysates made from wild-type control and srp102(K51I) cells during a time course after the temperature shift shows that the steady-state level of Srp102(K51I)p does not change significantly during the duration of the 10 h time course (Fig. 2 C, lanes 6–10). This result suggests that the temperature sensitivity of srp102(K51I) is not due to destabilization and subsequent degradation of Srp102(K51I)p at the restrictive temperature.
As an alternative means to deplete cells of Srp102p, we placed SRP102 under the control of the GAL1/GAL10 promoter to regulate its expression. Srp102p was depleted by switching cells from galactose-(where SRP102 is expressed) to glucose- (where SRP102 is repressed) containing media. We monitored growth and protein translocation as described above. As Srp102p was depleted, cells showed a fivefold decrease in their growth rate, and precursors of Kar2p and DPAP-B, but not CPY, accumulated to similar degrees as described above and shown previously in analogous experiments (Hann and Walter, 1991, Ogg et al., 1992, Brown et al., 1994) for the depletion of the other known components that function in the SRP- dependent protein targeting pathway (not shown).
Srp102p is Localized to the ER
To characterize the cellular disposition of Srp102p, we constructed two alleles in which the wildtype SRP102 gene is appended with sequences encoding epitopes that can be recognized by well-characterized monoclonal antibodies recognizing either a 12–amino acid sequence derived from influenza virus hemagglutinin (HA epitope; Field et al., 1988) or an 11–amino acid sequence derived from the mammalian c-myc protein (myc epitope; Evan et al., 1985). Both epitope-tagged versions—Srp102p-HA and Srp102p-myc—are fully functional (as judged by their ability to complement the growth defect of cells in which SRP102 is disrupted), and are specifically recognized by the cognate antibodies on Western blots (not shown).
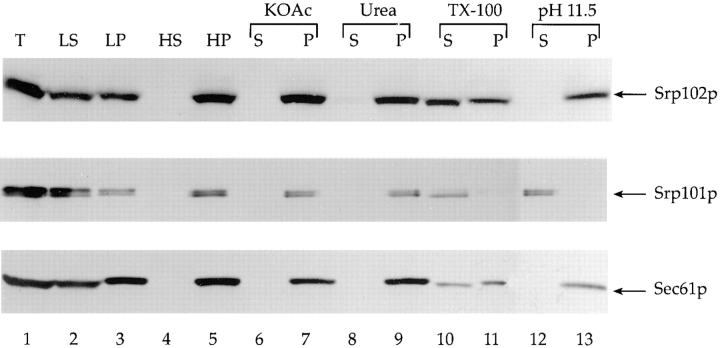
Based on its structural and functional homology to mammalian SRβ, Srp102p should be an integral membrane protein of the yeast ER. To test this prediction, we fractionated lysates prepared from cells expressing Srp102p-HA by differential centrifugation. After low-speed centrifugation, Srp102p-HA present in the total extract (Fig. 3, lane 1) partitioned about equally between the pellet (Fig. 3, lane 3) and the supernatant fractions (Fig. 3, lane 2). Presence of Srp102p in the low-speed pellet is presumably due to incomplete cell lysis. Further fractionation of the low-speed supernatant at 100,000 g resulted in complete sedimentation of Srp102p (Fig. 3, lanes 4 and 5), suggesting that Srp102p is associated with a particulate membrane fraction.
Figure 3.
Subcellular localization of Srp102p. Extracts from cells (srp102::HIS3 bearing pSO459) were fractionated as detailed in Materials and Methods. Equivalent amounts (0.5 OD600) of each fraction were analyzed by Western blot using anti-HA monoclonal antibodies, affinity-purified anti-Srp101p antibodies, or anti-Sec61p antibodies. Lane 1, total cell lysate(T); lane 2, low-speed supernatant (LS); lane 3, low-speed pellet (LP); lanes 6–13 contain the supernatant (lanes 6, 8, 10, and 12 [S]) or the pellet (lanes 7, 9, 11, and 13 [P]) after a portion of the low-speed supernatant was diluted with lysis buffer (lanes 4 and 5 [HS and HP]) or adjusted to a final concentrations of 600 mM potassium acetate (lanes 6 and 7 [KOAc]), 1.6 M urea (lanes 8 and 9 [Urea]), 0.5% Triton X-100 (lanes 10 and 11 [TX-100]), or 0.1 M Na2CO3 (lanes 12 and 13, [pH 11.5]) and centrifuged at 100,000 g. Only the relevant portion of each blot is shown. For comparison, the same fractions were also blotted with anti-Srp101p, a peripheral membrane protein, and anti-Sec61p, a previously characterized integral membrane protein of the ER.
The association of Srp102p with this fraction was further characterized by treatment of the low-speed supernatant with reagents known to perturb protein–protein or protein–lipid interactions before centrifugation at 100,000 g. Treatment with 600 mM potassium acetate (Fig. 3, lanes 6 and 7), 1.6 M urea (Fig. 3, lanes 8 and 9), or sodium carbonate at pH 11.5 (Fig. 3, lanes 12 and 13) did not solubilize Srp102p, which, under all conditions was recovered in the membrane fraction. In contrast, treating the low-speed supernatant with 0.5% Triton X-100 resulted in solubilization of roughly 50% of Srp102-HA, which was recovered in the high-speed supernatant fraction (Fig. 3, compare lanes 10 and 11).
As controls for these experiments, we also probed the Western blot with antibodies to Sec61p, a bona fide yeast ER integral membrane protein (Deshaies and Schekman, 1987), and Srp101p, which is a peripheral ER membrane protein (Ogg et al., 1992). Srp102p-HA cofractionated under all conditions with Sec61p. In particular, both Srp102p and Sec61p were recovered in the pellet fraction after alkali treatment, whereas Srp101p was completely extracted and recovered in the supernatant fraction (Fig. 3, compare lanes 12 and 13). Thus, we conclude that Srp102p-HA, and by extension Srp102p, is an integral membrane protein. This conclusion is consistent with the predictions from its amino acid sequence that shows a stretch of 19 hydrophobic amino acids forming a putative transmembrane domain near its amino terminus.
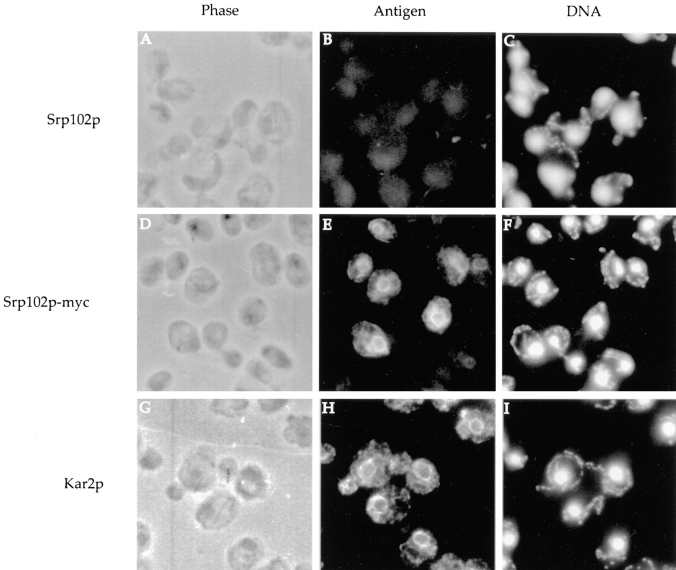
To further support our conclusion that Srp102p is a membrane protein of the yeast ER, we visualized the distribution of the epitope-tagged form of the protein by indirect immunofluorescence microscopy. Cells expressing the myc-tagged version of Srp102p from a 2μ plasmid are shown in Fig. 4. After formaldehyde fixation, cells were incubated with the appropriate primary and FITC, or TRITC-conjugated secondary antibodies (Fig. 4, B, E, and H); nuclei and mitochondria were visualized with the DNA-binding dye DAPI (Fig. 4, C, F, and I). Anti-myc antibodies detected the characteristic yeast ER-staining pattern (Fig. 4 E) consisting of a distinct perinuclear ring with fine strands of staining passing through the cytoplasm connecting the perinuclear ER to ER cisternae underlying the plasma membrane. Staining was variable; some cells in every field stained very brightly, while others stained less brightly or not at all. We attribute this staining to variation in the copy number of the 2μ plasmid in the population. Expression of Srp102p-HA from a CEN/ARS plasmid resulted in a less intense but qualitatively identical pattern (not shown). This pattern closely resembled that obtained in control samples stained with anti-Kar2p antibodies (Fig. 4 H). Wild-type cells (expressing Srp102p without an epitope tag) showed only background staining (Fig. 4 B). Taken together, the results from the fractionation experiments and the results from the immunofluorescence microscopy support our conjecture that Srp102p is an integral ER membrane protein.
Figure 4.
Immunofluorescent localization of Srp102p to the ER. SOY162 cells containing either pSO454 (Srp102p lacking an epitope tag, A–C and G–I) or pSO457 (Srp102p-myc, D–F) were fixed with formaldehyde and probed with anti-myc (A–F) or anti-Kar2p (G–I) followed by secondary decoration with FITC-conjugated donkey anti-mouse IgG (A–F) or TRITC-conjugated donkey anti–rabbit IgG (G–I). The image in B is exposed for twice the length of time as the image shown in E. Phase contrast images are shown in A, D, and G; fluorescein fluorescence is shown in B and E; rhodamine fluorescence is shown in H, and DAPI staining of the nuclei and mitochondria is shown in C, F, and I.
Srp101p and Srp102p Form a Stable Complex in the ER
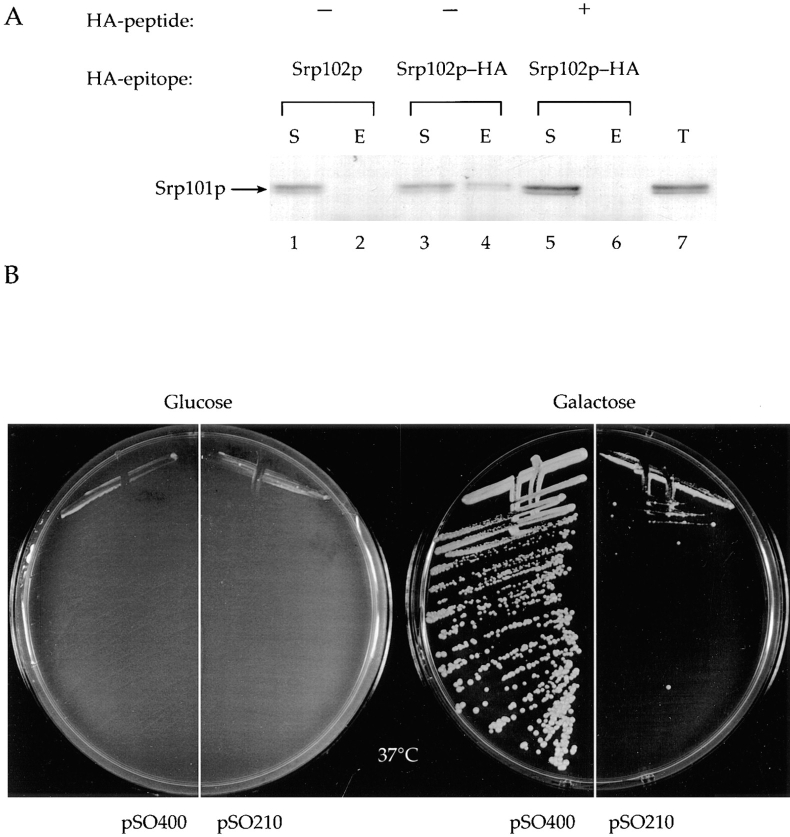
Mammalian SRα and SRβ are subunits of a heterodimeric complex (Tajima et al., 1986). To test whether the yeast homologues Srp101p and Srp102p are likewise associated, we asked if we could coimmunoprecipitate Srp101p with Srp102p. We immunoprecipitated Srp102p-HA with anti-HA antibodies from extracts prepared by detergent solubilization of yeast microsomal membranes under nondenaturing conditions. Immune complexes were subsequently solubilized in SDS sample buffer and subjected to SDS-PAGE followed by Western blot analysis. The blots were probed with affinity-purified polyclonal antibody directed against Srp101p. The results show that Srp101p was recovered in the eluate fraction from the HA-agarose beads when cells expressed Srp102p-HA (Fig. 5 A, lane 4), but was found exclusively in the supernatant fraction when cells expressed untagged Srp102p (Fig. 5 A, lanes 1 and 2) or when a peptide containing the HA epitope was added to compete for binding of the anti-HA antibody to Srp102p-HA (Fig. 5 A, lanes 5 and 6). These biochemical data strongly suggest that Srp101p and Srp102p directly interact to form a complex.
Figure 5.
Srp101p and Srp102p interact. (A) Detergent-solubilized microsomes were prepared from cells (lanes 1 and 2, SOY162 bearing pSO454, no tag; lanes 3–6, Srp102p-HA, SOY162 bearing pSO459) as described in Materials and Methods. Solubilized microsomes were subjected to immunoprecipitation using anti-HA either in the absence (lanes 1–4) or the presence (lanes 5 and 6) of excess HA peptide (NH2-YPYDVPDYA-COOH). Immunoprecipitated proteins were eluted from HA-agarose beads, subjected to SDS-PAGE, and analyzed by Western blot using affinity-purified anti-Srp101p antibodies. Eluates from each of the immunoprecipitations are shown in lanes 2, 4, and 6 (E); supernatants shown in lanes 1, 3, and 5 (S). Lane 7 contains 2 OD600 cell equivalents of solubilized membranes as a marker for the amount of Srp101p that is in each of the immunoprecipitations (T). (B) SOY162 cells bearing the srp102(K51I) allele on plasmid pSO462 were transformed with either pSO400 (SRP101 under galactose regulation) or pSO211 (SRP101 under control of its own promoter) and plated onto synthetic plates lacking tryptophan and uracil, and containing 2% galactose or 2% glucose as the sole carbon source. (Glucose), plate containing 2% glucose incubated at 37°C for 3 d. (Galactose) plate containing 2% galactose incubated at 37°C for 3 d.
In vivo evidence for an interaction between Srp101p and Srp102p was obtained by genetic means. To this end, we used cells expressing the temperature-sensitive srp102(K51I) allele. As temperature-sensitive mutations can often be suppressed by overproducing interacting proteins, we asked whether overexpression of Srp101p could suppress the growth defect induced at the restrictive temperature. We transformed cells expressing Srp102(K51I)p either with a plasmid bearing SRP101 under control of the GAL1/10 promoter, or with a plasmid bearing SRP101 under the control of its own promoter. Each of these strains grew at wild-type rates at the permissive temperature using either dextrose or galactose as the carbon source (not shown). As expected, at the restrictive temperature neither plasmid suppressed the growth defect when cells were grown on glucose-containing media (Fig. 5 B, glucose plate). In contrast, the plasmid bearing the galactose- inducible SRP101, but not the control SRP101 plasmid, suppressed the growth defect when cells were grown on galactose-containing media (Fig. 5 B, galactose plate). Under the galactose-induced conditions, Srp101p was ∼50-fold overexpressed as estimated by Western blot (Ogg et al., 1992). Significantly, galactose-inducible SRP101 failed to suppress the growth defect of cells containing a complete deletion of SRP102 rather than the srp(K51I) allele, even when these cells were grown in galactose (not shown).
Srp102p Anchors Srp101p to the ER Membrane
Biochemical studies that monitor extraction behavior of the SR subunits (both those shown in Fig. 3 and those performed in the mammalian system) suggested that SRα is a peripheral, whereas SRβ, which contains a TMD, is an integral membrane protein (Miller et al., 1995). This suggested a hierarchy of interactions with the membrane in which SRα binds to SRβ, which in turn is anchored in the membrane via its TMD. To test this notion in vivo, we asked whether Srp101p would be localized to the membrane even in the absence of Srp102p.
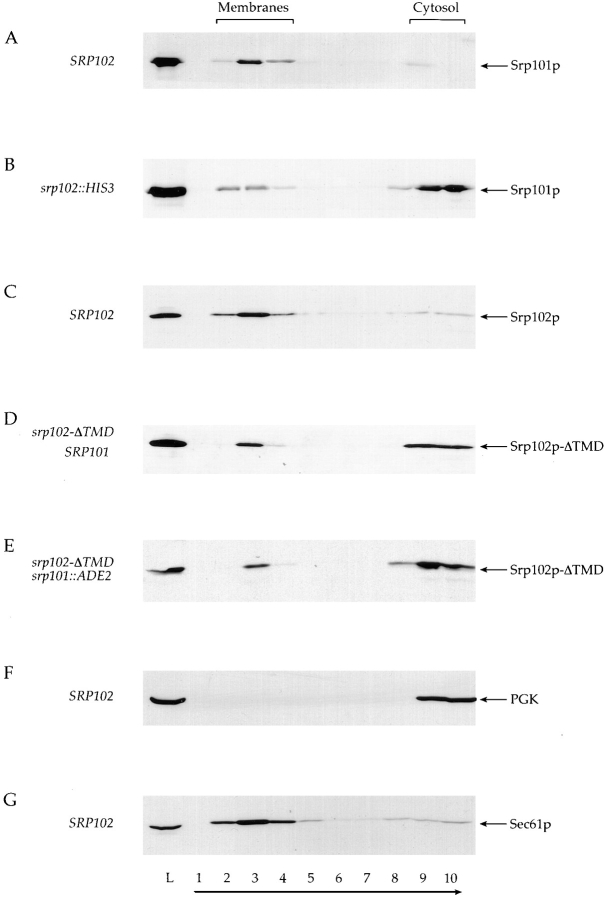
To this end, we prepared a crude cell extract as for the experiments described in Fig. 3, containing cellular membranes (including ER) as well as cytosol. To assess membrane association, we raised the density of the extract by adding sucrose, and placed this solution underneath a sucrose step gradient of lesser density (see Materials and Methods). Centrifugation of the gradient to equilibrium causes membranes to float to regions in the gradient that are equal to their buoyant density. In contrast to the differential velocity centrifugation used in Fig. 3, this method allows us to distinguish conclusively the membrane association of components (in which case they will float to regions of lighter density) from aggregation (in which case they will remain in the denser sucrose fractions or sediment into a pellet). After centrifugation, phosphoglucokinase (PGK), a cytosolic enzyme marker, consistently remained in fractions 9 and 10 at the bottom of the gradient (Fig. 6 F), while most of Sec61p, an integral ER membrane protein, moved to fractions 2–4 (Fig. 6 G), thereby identifying these fractions as those that contain the ER.
Figure 6.
Fractionation behavior of Srp101p and Srp102 during perturbation of the SR. Whole-cell lysate from cells was layered under a 2.4-ml discontinuous sucrose gradient, and was centrifuged to equilibrium. Equivalent amounts of the load fraction (lane L) and fractions from the gradient (lanes 1–10) were analyzed by Western blot using anti-Srp101p, anti-Srp102p, anti-PGK, or anti-Sec61p antisera. Fractionation behavior of Srp101p was compared between strains containing either wild-type Srp102p ([A] SRP102, WBY618 bearing pSO452) or the srp102:: HIS3 deletion ([B], WBY618). As controls, fractionation of the cytosolic enzyme phosphoglucokinase ([F] PGK) or the integral ER membrane protein Sec61p (G) was also monitored in a strain wild-type for SRP102 (WBY618 bearing pSO452) to identify those fractions containing cytosol or membranes. Srp102p fractionation was also monitored in wild-type cells ([C] SRP102, WBY618 bearing pSO452) as was fractionation of Srp102p-ΔTMD in cells either containing or lacking Srp101p ([D] WBY618 bearing pWB209; [E] WBY823). Direction of sedimentation is indicated by an arrow, and the fractions containing cytosol (lanes 9 and 10) or membranes (lanes 2–4) are also indicated above.
As expected, the majority of Srp101p was found associated with membranes (Fig. 6 A, compare fractions 2–4 with fractions 9 and 10) when we monitored the fractionation behavior of the protein in extracts prepared from wild-type cells (SRP102). This ratio is reversed in srp102:: HIS3 cells (Fig. 6 B). Thus, deletion of Srp102p resulted in the release of the majority of Srp101p into the cytosol. A significant fraction of Srp101p, however, still cofractionated with membranes from cells in which Srp102p was deleted. In contrast, the vast majority of Srp102p was always found in the membrane fractions, irrespective of whether SRP101 was deleted or not (data not shown). From these data we conclude that Srp102p contributes significantly to the localization of SR to the ER membrane. A minor fraction of Srp101p, however, still interacts (directly or indirectly) with membranes, even in the absence of its partner Srp102p. As srp102 mutant cells are phenotypically identical to mutant cells lacking other SRP pathway components (see above); however, the small amount of Srp101p that is membrane-associated in a srp102::HIS3 cell is not functional, or is insufficient to provide significant SR activity.
Mutations in the GTP-binding Site Result in Compromised SR Complex Formation
Much of the cytosolic portion of Srp102p constitutes its GTPase domain. We therefore considered the possibility that the GTPase domain of Srp102p might play a role in anchoring Srp101p. We made site-directed mutations in the predicted GTP-binding site of Srp102p, in particular in the consensus motifs (G-boxes) that by analogy to other GTPases are predicted to constitute loops on the surface of the domain that directly contribute to the nucleotide-binding pocket (Bourne et al., 1991). Many mutations in these conserved elements of other GTPases have been made and functionally characterized. In many cases, corresponding mutations in related GTPases have similar consequences on the enzymatic properties of the enzymes, allowing the molecular phenotypes to be predicted. Based on the information available for other GTPases, we made site-directed point mutations in Srp102p based on their predicted effect (listed in Table II together with predicted molecular phenotypes, if known). In addition to nine single amino acid changes, we made several mutations that either changed multiple amino acids in a single G-box, or changed amino acids in multiple G-boxes at the same time (Table II).
Table II.
Site-directed Mutations in the GTP Binding Site of Srp102p
| Allele | G-box location | Expression | Phenotype | Effect of mutation | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Single amino acid changes in a single G-box | ||||||||||
| S49A | G-1 | Yes | wild type | reduced GTP hydrolysis | Barbacid, 1987 | |||||
| K51I | G-1 | Yes | ts | reduced nucleotide affinity | Pai et al., 1990 | |||||
| T52N | G-1 | Yes | ts | increased affinity for GEF, | ||||||
| reduced affinity for GTP | Peyroche et al., 1996 | |||||||||
| T66A | G-2 | Yes | wild type | prevents GTP-dependent | ||||||
| interaction with GAP | Sigal et al., 1986 | |||||||||
| G90L | G-3 | Yes | ts | unknown | ||||||
| H91L | G-3 | Yes | wild type | reduced GTPase activity | Jonak et al., 1994 | |||||
| N154I | G-4 | Yes | ts | impaired nucleotide exchange | Sigal et al., 1986 | |||||
| E157Q | G-4 | Yes | wild type | reduced affinity for GTP | Weijland et al., 1994 | |||||
| S220A | G-5 | ND | wild type | bypass requirement for GEF | Camonis and Jacquet, 1988 | |||||
| Multiple amino acid changes in a single G-box | ||||||||||
| P46A,Q47A,N48A,ΔS49 | G-1 | Yes | wild type | unknown | ||||||
| K51A,T52A | G-1 | Yes | wild type | unknown | ||||||
| N154A,E157A | G-4 | Yes | wild type | unknown | ||||||
| Multiple amino acid changes in multiple G-boxes | ||||||||||
| H91L,N154I | G-3, G-4 | Yes | ts | unknown | ||||||
| H91L,N154A,E157A | G-3, G-4 | ND | ts | unknown | ||||||
| H91L,E157Q | G-3, G-4 | Yes | ts | unknown | ||||||
| K51I,H91L | G-1, G-3 | Yes | null | unknown | ||||||
| K51I,N154I | G-1, G-4 | ND | null | unknown | ||||||
| K51A,T52A,H91L | G-1, G-3 | Yes | null | unknown | ||||||
| K51A,T52A,E157Q | G-1, G-4 | ND | null | unknown | ||||||
Each site-directed mutation is listed along with its location in G-boxes 1–5 (nomenclature according to Bourne et al., 1991) and the observed growth phenotype when the mutant allele is present as the only copy of Srp102p is a haploid cell. Except for the mutants indicated by ND, wild-type expression levels of mutant proteins were confirmed by Western blot analysis. Most mutations were chosen on the basis of their predicted molecular phenotypes inferred from characterized defects in other GTPases as indicated.
Plasmids encoding the mutant forms of SRP102 were transformed into diploid SRP102/srp102::HIS3 cells, which were then sporulated, and haploid cells bearing both the chromosomal srp102::HIS3 deletion and the plasmid were selected. Using this approach rather than transforming a haploid SRP102–deleted strain directly, we avoided potential complication arising from cells having grown in the absence of the SRP-dependent targeting pathway, such as their turning rho−. Table II lists the growth phenotype of each mutation. Surprisingly, in eight of the twelve cases where mutations are confined to a single G-box, cells carrying the mutant allele grew with wild-type rates. The other four mutants (K51I, T52N, G90L, and N154I) resulted in cells that were temperature-sensitive for growth and for protein translocation (the results for K51I were discussed above in Fig. 2, A and B). In contrast, in each of the seven cases where an allele consisted of mutations in more than one of the G-boxes, we observed either a temperature-sensitive or null phenotype (Table II), indicating that mutations in more than one of the G-boxes are far more detrimental than the same mutations in either one of the boxes. For example, combination of the K51A/T52A allele with the H91L allele resulted in a null phenotype, whereas no growth phenotype was observed for either of those mutations by themselves.
Western blots revealed that even when combinatorial mutants result in a null phenotype, the mutant forms of Srp102p were still present in cells at wild-type levels. Fig. 2 C, for example, shows that srp102(K51I) does not cause Srp102p to be degraded at the nonpermissive temperature. Similar results were obtained for the other mutant alleles (Table II). Thus, the mutant phenotypes are not due to degradation of the mutant forms of Srp102p.
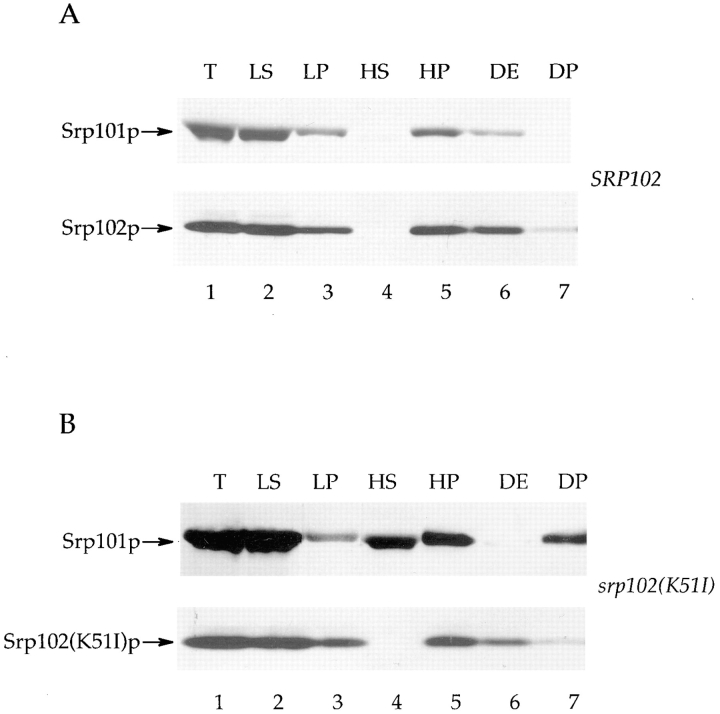
To gain further insight into the biochemical defect causing the temperature sensitivity of srp102(K51I) cells, we fractionated extracts prepared from cells expressing an HA-tagged version of the mutant protein (Srp102p[K51I] p-HA) by differential centrifugation. Fig. 7 shows the results of such an experiment in which cells had been grown at the permissive temperature 24°C. As expected and recapitulating the results shown in Fig. 3, Srp101p and Srp102p cofractionate in SRP102 wild-type cells (Fig. 7 A). In srp102(K51I) cells the fractionation of Srp102p is unchanged, but strikingly, the fractionation of Srp101p is grossly altered (Fig. 7 B). In particular, about half of the Srp101p that is present in the low-speed supernatant fraction (lane 2) is now recovered as an apparently soluble protein in a high-speed supernatant (lane 4). Moreover, the remaining portion that partitioned into the high-speed pellet fraction was not extracted by detergent (1% Triton X-100), but instead was recovered as an apparent aggregate in the pellet fraction (lane 7). From these studies we conclude that the interaction between Srp101p and Srp102p is severely altered in srp102(K51I) mutant cells, even at permissive conditions, and that increased temperature exacerbates this defect, leading to the temperature-sensitive phenotype. In support of this hypothesis is the fact that overexpression of Srp101p in srp102(K51I) mutant cells rescues the growth defect seen at the nonpermissive temperature in these cells (Fig. 5 B).
Figure 7.
SR subunits dissociate in cells bearing srp102(K51I)p. Cells bearing wild-type Srp102p-HA (SOY162 containing pSO459) or Srp102(K51I)p-HA (SOY162 containing pSO462) were subjected to fractionation as detailed in Materials and Methods. Equivalent amounts of each fraction were subjected to SDS-PAGE, followed by Western blot analysis. Srp101p was detected with affinity-purified anti-Srp101p antibodies, while Srp102p was detected using the anti-HA monoclonal antibody. Each panel contains the fractions loaded in the same manner. Lane 1: (T) total cell lysate; lane 2: (LS), low-speed supernatant; lane 3 (LP), low-speed pellet; lane 4, (HS), high-speed supernatant; lane 5, (HP), high-speed pellet; lane 6, (DE), detergent extract; lane 7, (DP), detergent pellet. (A) fractions from Srp102p-HA cells grown at 24°C. (B) fractions from Srp102(K51I)p-HA grown at 24°C.
We also tested four additional temperature-sensitive mutant strains for suppression by the Srp101p-overexpressing plasmid pSO400. The growth defects at the nonpermissive temperature of three of these strains, bearing srp102(G90L), srp102(N154I), and srp102(H91L, E157Q), respectively, were all suppressed upon overproduction of Srp101p (T. Hu and P. Walter, unpublished observation). This result suggests that the observed effect is not allele-specific, but that, as for srp102(K51I) cells, the affinity of Srp102p and Srp101p may be weakened by the mutations. In contrast, srp102(T52N) cells remained temperature-sensitive, even when Srp101p was overexpressed; this mutation therefore defines a phenotypically distinct class of mutants.
Deletion of the Transmembrane Domain of SRP102 Does Not Perturb its Function
The results presented so far suggested that the GTPase domain of Srp102p may be involved in recruiting Srp101p to the ER membrane and, in this role, may be essential for SRP receptor function. We decided to test this notion by perturbing the membrane association of the SRP receptor by a different strategy. To this end, we constructed an allele of SRP102 in which the 19–amino acid TMD was deleted. We expected that srp102-ΔTMD cells would exhibit the phenotype characteristic of cells lacking the SRP- dependent protein targeting pathway, because neither Srp101p nor Srp102p would be localized correctly to the ER membrane. To our surprise, we found that the srp102-ΔTMD mutant allele efficiently complemented the growth defect of srp102::HIS3 disruption cells. As shown in Fig. 1, the growth rate of cells expressing Srp102p-ΔTMD resembles the growth rate of cells expressing wild-type Srp102p, and is clearly distinct from the growth rate seen in cells containing the SRP102 disruption.
These results suggested that Srp102p-ΔTMD may be capable of performing its function in SRP-dependent protein targeting with close to wild-type efficiency. To test this notion further, we asked whether the Srp102p-ΔTMD allele compromised protein translocation in vivo. We pulse- labeled Srp102p-ΔTMD cells with [35S]methionine, and immunoprecipitated proteins known to be translocated into the ER as described for Fig. 2. Neither substrates that are SRP-dependent (e.g., DPAP-B) nor substrates that are SRP-independent (e.g. CPY) showed detectable translocation defects (not shown). Thus, we conclude that the TMD of Srp102p is not required for SR to perform its function.
To test whether removal of the TMD releases Srp102p from the ER membrane, we determined the intracellular distribution of Srp102p-ΔTMD by subcellular fractionation and indirect immunofluorescence. To this end, we subjected a lysate derived from cells expressing Srp102p-ΔTMD to isopycnic centrifugation in a sucrose step gradient. As shown in Fig. 6, the majority of Srp102p-ΔTMD indeed remains at the bottom of the gradient in contrast to Srp102p found in wild-type cells (compare Fig. 6 D, fractions 9 and 10, with Fig. 6 C, fractions 9 and 10), whereas most of the ER membrane in the mutant cells floated to fractions 3 and 4 as indicated by fractionation of the Sec61p integral membrane marker protein (not shown). The primarily cytoplasmic localization of Srp102-ΔTMD was confirmed by indirect immunofluorescence (not shown). A small portion (∼25%) of Srp102p-ΔTMD did, however, cofractionate with ER membrane (Fig. 6 D, lane 3).
Because Srp101p may contain additional residues that contribute to association of the SR with the ER membrane (Hortsch et al., 1985; Lauffer et al., 1985), we next asked whether we could achieve a complete release of Srp102p-ΔTMD into cytosolic fractions in a cell in which both Srp101p and the TMD of Srp102p were deleted. As shown in Fig. 6 E, the fractionation behavior of Srp102p-ΔTMD is identical whether or not Srp101p is present in the cells. From these data, we conclude that Srp102p is likely to interact with ER membranes through interactions in addition to those afforded by its TMD or its partner subunit Srp101p.
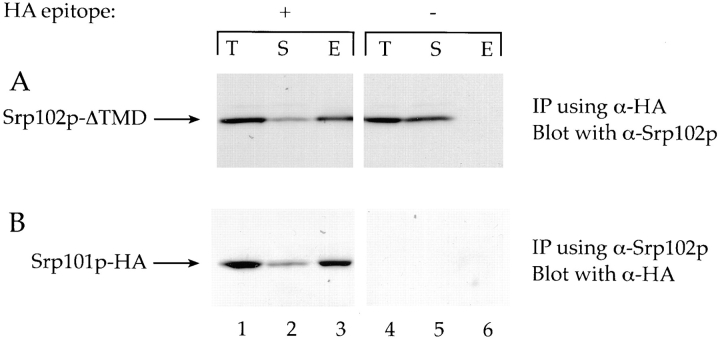
Because deletion of the TMD of Srp102p showed little effect on cell growth or on protein translocation, and because the data presented above suggest that the majority of Srp102p-ΔTMD has been released from the ER membrane, we asked whether Srp101p and Srp102p-ΔTMD still form a heterodimer in the cytosol. To this end, we prepared a cytosol fraction from Srp102p-ΔTMD cells (also bearing an HA-tagged allele of SRP101) and performed a coimmunoprecipitation experiment similar to that shown in Fig. 5. Coimmunoprecipitated proteins were separated by SDS-PAGE; Srp101p, and Srp102p-ΔTMD were detected by Western blotting using polyclonal rabbit antisera (Fig. 8). A significant portion of Srp102p-ΔTMD in the extract (Fig. 8 A, lane 1, T) was recovered in the eluate fraction from the HA-agarose beads when cells expressed the HA-tagged version of Srp101p (Fig. 8 A, lane 3, E), but was found exclusively in the supernatant fraction when cells contain untagged Srp101p (Fig. 8 A, lane 5, S), indicating that Srp102p-ΔTMD is associated with Srp101p-HA in the extract. This result was confirmed by the converse experiment: anti-Srp102p serum immunoprecipitated Srp101p-HA (Fig. 8 B, lane 3), followed by detection of Srp101p-HA with the monoclonal antibody directed against the HA tag.
Figure 8.
Srp102p-ΔTMD and Srp101p form a stable complex in the cytosol. A cytosolic extract was prepared from strains containing Srp102p-ΔTMD and Srp101p with or without an HA tag (lanes 1–3, Srp101p-HA, WBY752; anes 4– 6, no tag; WBY618 with pWB209). In A, 1 OD600 equivalent of cell cytosol was subjected to immunoprecipitation with anti-HA mAbs coupled directly to agarose beads. Coimmunoprecipitated proteins were eluted from the beads, and were then subjected to SDS-PAGE followed by Western blot analysis using antisera against Srp102p. Eluates from the immunoprecipitation are shown in lanes 3 and 6 (E, eluate), while proteins not bound during the immunoprecipitation are shown in lanes 2 and 5 (S, supernatant). Lanes 1 and 4 (T, total) contain 1 OD600 cell equivalent of whole-cell extract as a marker for the amount of Srp102p-ΔTMD that is in each of the immunoprecipitations. B shows the same type of experiment, but using the anti-Srp102p antibody coupled to beads to perform the immunoprecipitation followed by detection of Srp101p-HA with anti-HA mAbs. Note that lanes 4–6 are blank because the Srp101p present in those lanes has no HA epitope.
Discussion
Our results demonstrate that Srp102p is the yeast homologue of SRβ. This notion is strongly supported by several independent lines of evidence: (a) by Srp102p's strong sequence and structural similarity to the mammalian SRβ; (b) by its disposition as an integral, ER-localized membrane protein; (c) by the growth and protein translocation defects exhibited in srp102 mutant cells, and (d) by its physical association and genetic interaction with Srp101p. Like mammalian SRβ, Srp102p is a GTPase that is anchored by an amino-terminal TMD in the ER membrane. The identification of the yeast SRβ has allowed us to test experimentally the role of its GTP binding site and its TMD. Both lines of investigation yielded rather unexpected results. First, the GTP binding site of Srp102p appears to be surprisingly resilient to mutations that inactivate other GTPases. When the GTP binding site is sufficiently disturbed, however, SR function is lost. Interestingly, in most cases tested the loss of activity parallels a loss of the affinity of Srp102p for Srp101p. Second and equally surprising, we find that the TMD of Srp102p is not required for its function. Thus, the SR can function as a soluble cytosolic factor that may only associate transiently with ER membrane components most likely through an interaction with other membrane proteins.
To our knowledge, the relative insensitivity of the GTP binding site of Srp102p to mutational perturbation is unprecedented by studies of other GTPases. Mutations were chosen based on their predicted effects on blocking different steps in the presumptive Srp102p GTPase cycle; yet eight of the twelve mutations that change single or multiple amino acids in single G-boxes in Srp102p have no discernible effect on the growth of cells or on SRP-dependent protein translocation. We consider it likely that these mutations have not caused enough of a change in steady-state levels of the different nucleotide-bound forms of Srp102p to exhibit a mutant phenotype. In each case tested, however, combination of mutations in single G boxes such that now residues of two separate G-boxes are altered, resulted in more severely impaired SR function. In all cases analyzed, the expression levels of Srp102p mutants were not affected, thus making it unlikely that combination of mutations caused global unfolding (which might be expected to destabilize the protein in cells). We envision that each G-box loop on the GTPase domain contributes binding energy through contacts with the nucleotide. Upon disruption of the contributions of a single loop, nucleotide binding is still sufficiently tight to support Srp102p function, whereas perturbation of multiple loops lowers the affinity below a required threshold. It follows from this interpretation that none of the mutated side chains can be absolutely required for Srp102p's catalytic activity, i.e., each of the mutant Srp102ps must still be capable of traversing SRβ's GTPase cycle.
SRP-dependent protein targeting is evolutionarily conserved. In bacteria, homologues of SRP54 and SRα (termed Ffh and FtsY, respectively) have been found to play an important role in membrane protein biogenesis (Seluanov and Bibi, 1997; Ulbrandt et al., 1997). In contrast, no recognizable homologue for SRβ is encoded in the bacterial genome; SRβ therefore may be an exclusively eukaryotic invention. Our results show that the yeast SR does not have to be an integral membrane protein to be functional; thus, the role of SRβ is unlikely to be solely that of a membrane anchor for SRα. Rather, our results suggest that an interaction between SRα and SRβ is required for targeting. As borne out by the mutational analysis, this interaction in turn is dependent on features in the GTPase domain of Srp102p, possibly reflecting regulation by its nucleotide-bound state. Thus, SRβ could function as an enabling switch that is required to recruit SRα from an inactive pool into a heterodimeric complex that then is active to promote protein targeting. Prokaryotic FtsY may be constitutively active, not requiring such a switch. In this role, SRβ might increase the fidelity of protein targeting by sensing the status of downstream components (e.g., the translocon) and activate SRα only when unoccupied translocation sites are available. The interaction of the activated SRα with the SRP/ribosome/nascent chain complex may then carry out the nuts-and-bolts reaction that releases the signal sequence from SRP and ascertains proper docking of the ribosome onto the translocon.
Clearly, other models remain possible to account for the presented results. To decipher the role of the GTPase domain of SRβ, biochemical analyses will be required to extend the mutational studies initiated here and to define the role of individual nucleotide-bound states of Srp102p. The availability of active, soluble Srp101p–Srp102p-ΔTMD complexes should greatly facilitate such analyses, as this will allow functional studies without a requirement for tedious and often inefficient reconstitutions of detergent solubilized SRP receptor into artificial lipid bilayers.
Acknowledgments
We wish to thank Tianhua Hu, Ted Powers, Gustavo Pesce, and members of the Walter laboratory for discussions and comments on the manuscript. We thank Caroline Shamu, Peter Sorger, Tim Stearns, and Joe Vogel for strains and plasmids.
This work was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft to W.P. Barz, a Howard Hughes predoctoral fellowship to S.C. Ogg, and by grants from the National Institutes of Health. P. Walter is an Investigator of the Howard Hughes Medical Institute.
Abbreviations used in this paper
- CPY
carboxypeptidase Y
- DPAP-B
dipeptidyl aminopeptidase B
- FB
flotation buffer
- SR
SRP receptor
- SRα
SRP receptor α-subunit
- SRβ
SRP receptor β-subunit
- SRP
signal recognition particle
- TMD
transmembrane domain
Footnotes
The current addresses of Stephen C. Ogg is Department of Biochemistry, University of Dundee, Dundee DD1 4HN, Scotland. The current address of Wolfgang P. Barz is Department of Membrane Biochemistry, Max-Planck-Institute for Biochemistry, D-82152 Martinsried, Germany.
Address all correspondence to Peter Walter, Howard Hughes Medical Institute and Department of Biochemistry and Biophysics, University of California School of Medicine, San Francisco, CA 94143-0448. Tel.: 415-476-5017; Fax: 415-476-5233; E-mail: walter@cgl.ucsf.edu
References
- Bacher G, Lütcke H, Jungnickel B, Rapoport TA, Dobberstein B. Regulation by the ribosome of the GTPase cycle of the signal-recognition particle during protein targeting. Nature. 1996;381:248–251. doi: 10.1038/381248a0. [DOI] [PubMed] [Google Scholar]
- Barbacid M. rasGenes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54k subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Brown JD, Hann BC, Medzihradszky KF, Niwa M, Burlingame AL, Walter P. Subunits of the Saccharomyces cerevisiaesignal recognition particle required for its functional expression. EMBO (Eur Mol Biol Organ) J. 1994;13:4390–4400. doi: 10.1002/j.1460-2075.1994.tb06759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Ng DTW, Ogg SC, Walter P. Targeting Pathways to the Endoplasmic Reticulum Membrane. Cold Spring Harb Symp Quant Biol. 1995;60:23–30. doi: 10.1101/sqb.1995.060.01.005. [DOI] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. . Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Camonis JH, Jacquet M. A new RAS mutation that suppresses the CDC25 gene requirement for growth of Saccharomyces cerevisiae. . Mol Cell Biol. 1988;8:2980–2983. doi: 10.1128/mcb.8.7.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Connolly T, Gilmore R. Formation of a functional ribosome–membrane junction during translocation requires the participation of a GTP-binding protein. J Cell Biol. 1986;103:2253–2261. doi: 10.1083/jcb.103.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Gilmore R. The signal recognition particle receptor mediates the GTP-dependent displacement of SRP from the signal sequence of the nascent polypeptide. Cell. 1989;57:599–610. doi: 10.1016/0092-8674(89)90129-3. [DOI] [PubMed] [Google Scholar]
- Davis RW, Thomas M, Cameron J, St. John TP, Scherer S, Padgett RA. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymology. 1980;65:404–406. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J Cell Biol. 1987;105:633–645. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. Structural and functional dissection of Sec62p, a membrane-bound component of the yeast endoplasmic reticulum protein import machinery. Mol Cell Biol. 1990;10:6024–6035. doi: 10.1128/mcb.10.11.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-mycproto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson IA, Lerner RA, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiaeby use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- Geitz RD, Sugino A. New yeast-Escherichia colishuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gilmore R, Blobel G, Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982a;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982b;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Hanein D, Matlack KE, Jungnickel B, Plath K, Kalies KU, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- Hann BC, Walter P. The Signal Recognition Particle in S. cerevisiae. . Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortsch M, Avossa D, Meyer DI. A structural and functional analysis of the docking protein. J Biol Chem. 1985;260:9137–9145. [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak J, Anborgh PH, Parmeggiani A. Histidine-118 of elongation factor Tu: its role in aminoacyl-tRNA binding and regulation of the GTPase activity. FEBS Lett. 1994;343:94–98. doi: 10.1016/0014-5793(94)80614-4. [DOI] [PubMed] [Google Scholar]
- Krieg UC, Walter P, Johnson AE. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc Natl Acad Sci USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kurzchalia TV, Wiedmann M, Girshovich AS, Bochkareva ES, Bielka H, Rapoport TA. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- Lauffer L, Garcia PD, Harkins RN, Coussens L, Ullrich A, Walter P. Topology of the SRP receptor in the endoplasmic reticulum membrane. Nature. 1985;318:334–338. doi: 10.1038/318334a0. [DOI] [PubMed] [Google Scholar]
- Meyer DI, Louvard D, Dobberstein B. Characterization of molecules involved in protein translocation using a specific antibody. J Cell Biol. 1982;92:579–583. doi: 10.1083/jcb.92.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, Tajima S, Lauffer L, Walter P. The β-subunit of the signal recognition particle receptor is a transmembrane GTPase that anchors the α-subunit, a peripheral membrane GTPase, to the endoplasmic reticulum membrane. J Cell Biol. 1995;128:273–282. doi: 10.1083/jcb.128.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, Wilhelm H, Gierasch L, Gilmore R, Walter P. GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature. 1993;366:351–354. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- Montoya G, Svensson C, Luirink J, Sinning I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 1997;385:365–368. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- Ng DTW, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Poritz M, Walter P. The signal recognition particle receptor is important for growth and protein secretion in Saccharomyces cerevisiae. . Mol Biol Cell. 1992;3:895–911. doi: 10.1091/mbc.3.8.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Walter P. Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science. 1995;269:1422–1424. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:358–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Rapiejko PJ, Gilmore R. Empty site forms of the SRP54 and SR alpha GTPases mediate targeting of ribosome-nascent chain complexes to the endoplasmic reticulum. Cell. 1997;89:703–713. doi: 10.1016/s0092-8674(00)80253-6. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Römisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B. Homology of the 54K Protein of signal recognition particle, docking protein, and two E. coliproteins with putative GTP-binding domains. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.M. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Plainview, New York. 545 pp.
- Seluanov A, Bibi E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- Sherman, F., and C. Lawrence. 1974. Saccharomyces. In Handbook of Genetics. R.C. King, editor. Plenum Press, New York. 359–393.
- Siegel V, Walter P. Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell. 1988;52:39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Heiter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia colias fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Tajima S, Lauffer L, Rath VL, Walter P. The signal recognition particle receptor is a complex that contains two distinct polypeptide chains. J Cell Biol. 1986;103:1167–1178. doi: 10.1083/jcb.103.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrandt ND, Newitt JA, Bernstein HD. The E. colisignal recognition particle is required of the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G. Purification of membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci USA. 1980;77:7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Zopf D, Bernstein HD, Johnson AE, Walter P. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO (Eur Mol Biol Organ) J. 1990;9:4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]