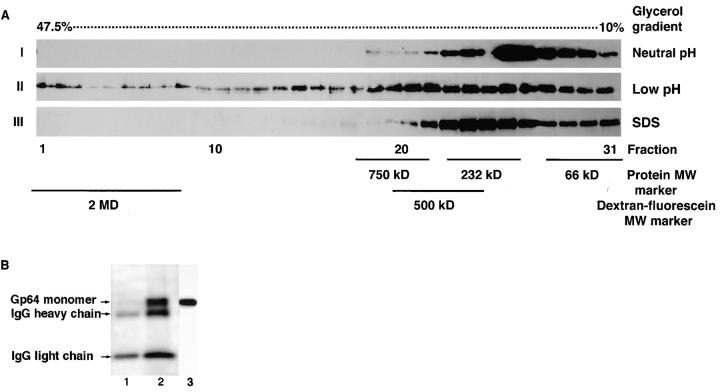
Figure 7.
Gp64 HC were formed in response to low pH activation. (A) Sf9 cells infected with baculovirus at m.o.i. of 1 were treated with neutral (I) or pH 4.9 (II and III) medium for 5 min. After this treatment, cell–surface cross-linking with DTSSP was performed to arrest gp64 protein complexes. Cross-linked cells were lysed in 1.5% Triton X-100 (I and II) or SDS (III) lysis buffer and subjected to 10– 47.5% glycerol density gradient ultracentrifugation. 31 fractions were collected and a 75-μl aliquot of each fraction was reduced with 50 mM DTT and analyzed in 10% SDS-PAGE. Reduction of each fraction released monomers from the cross-linked protein complex. External protein molecular weight standards (i.e., 66 kD albumin, 232 kD catalase, and 750 kD macroglobulin) and fluorescein-conjugated dextran (500 kD and 2 MD) were run in parallel with the test samples to estimate the MW of the cross-linked protein complexes. Replacing Triton X-100 by 1.5% SDS in III resulted in disappearance of the gp64 HC. (B) To determine whether besides gp64 our protein complex contains other unrelated proteins, infected Sf9 cells were cell–surface biotinylated, treated with pH 7.4 medium (lane 1) or pH 4.9 (lane 2) for 5 min, and then cross-linked with 5 mM DTSSP. Lysis was performed in 1.5% Triton X-100 followed by glycerol density gradient ultracentrifugation. The 10 heaviest fractions from glycerol density gradient were pooled and AcV1 mAb-immunopurified gp64 was analyzed in 10% SDS-PAGE under reducing conditions. Biotinylated proteins were detected with streptavidin (1:3,000) conjugated to horseradish peroxidase. Lane 3 represents unimmunopurified monomer control detected by AcV5 Mab (1:100) ran in parallel with biotinylated samples.