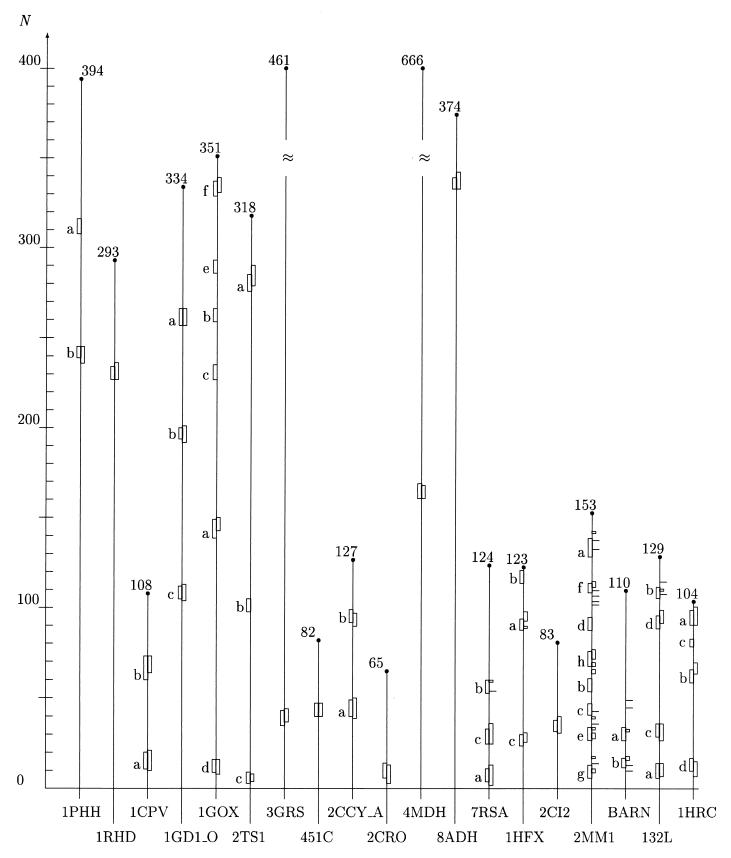
Figure 3.
Initiation sites of folding assuming helical structure determined by means of the minimal entropy criterion and other methods. On the abscissa, we have listed the Brookhaven codes of the proteins examined. The grid on the N-axis marks the position of the amino acids along the protein backbones, which have been skeletonized as vertical filaments of appropriate length. For convenience, the total number of residues of each protein is shown at the upper end of each filament. The boxes attached to each protein backbone highlight the location and extent of the earliest stretches of helical structure being formed during the folding process. The boxes on the left side of each filament have been positioned in the regions around the lowest minima of the average entropy 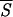 5 according to the criterion described in the caption of Fig. 1. The boxes or tick marks (in the case of single residues) on the right side of each backbone visualize the initiation sites as they result from experimental and computational works. Our segments are labeled with letters indicating ascending values of the entropy minima. The data for the first 12 proteins (1PHH, 1RHD, 1CPV, 1GD1
5 according to the criterion described in the caption of Fig. 1. The boxes or tick marks (in the case of single residues) on the right side of each backbone visualize the initiation sites as they result from experimental and computational works. Our segments are labeled with letters indicating ascending values of the entropy minima. The data for the first 12 proteins (1PHH, 1RHD, 1CPV, 1GD1 O, 1GOX, 2TS1, 3GRS, 451C, 2CCY
O, 1GOX, 2TS1, 3GRS, 451C, 2CCY A, 2CRO, 4MDH, and 8ADH) are drawn from ref. 49; the experimental results for 7RSA have been drawn from ref. 11 for site a and c and ref. 62 for site b. The site a also has been predicted in ref. 12 in the same position as determined in ref. 11. Helix c (but the last two residues) also has been shown to be present in the early folding intermediate identified in ref. 62. The experimentally detected sites for 1HFX, 2CI2, 2MM1, BARN (barnase), 132L, and 1HRC are drawn, respectively, from refs. 35, 60, 3, 61, 34, and 64. Our initiation site for 2CI2 includes the first two residues that comprise the folding nucleus according to ref. 65. The experimental data reported in ref. 3 for 2MM1 and ref. 34 for 132L essentially confirm the previous analysis in refs. 13 and 63, respectively. In the following, we list the range of variation δS of the entropy in the entropy profile and the site entropy S for each protein examined. 1PHH: δS = 0.224–0.690, S(a) = 0.224, S(b) = 0.253; 1RHD: δS = 0.173–0.687, S=0.173; 1CPV: δS=0.237–0.677, S(a) = 0.237, S(b) = 0.305; 1GD1
A, 2CRO, 4MDH, and 8ADH) are drawn from ref. 49; the experimental results for 7RSA have been drawn from ref. 11 for site a and c and ref. 62 for site b. The site a also has been predicted in ref. 12 in the same position as determined in ref. 11. Helix c (but the last two residues) also has been shown to be present in the early folding intermediate identified in ref. 62. The experimentally detected sites for 1HFX, 2CI2, 2MM1, BARN (barnase), 132L, and 1HRC are drawn, respectively, from refs. 35, 60, 3, 61, 34, and 64. Our initiation site for 2CI2 includes the first two residues that comprise the folding nucleus according to ref. 65. The experimental data reported in ref. 3 for 2MM1 and ref. 34 for 132L essentially confirm the previous analysis in refs. 13 and 63, respectively. In the following, we list the range of variation δS of the entropy in the entropy profile and the site entropy S for each protein examined. 1PHH: δS = 0.224–0.690, S(a) = 0.224, S(b) = 0.253; 1RHD: δS = 0.173–0.687, S=0.173; 1CPV: δS=0.237–0.677, S(a) = 0.237, S(b) = 0.305; 1GD1 O: δS = 0.125–0.689, S(a) = 0.125, S(b) = 0.166, S(c) = 0.192; 1GOX: δS = 0.288–0.691, S(a) = 0.288, S(b) = 0.290, S(c) = 0.294, S(d) = 0.307, S(e) = 0.311, S(f) = 0.315; 2TS1: δS = 0.227–0.687, S(a) = 0.256, S(b) = 0.273, S(c) = 0.284; 3GRS: δS = 0.154–0.688, S = 0.245; 451C: δS = 0.142–0.688, S = 0.237; 2CCY
O: δS = 0.125–0.689, S(a) = 0.125, S(b) = 0.166, S(c) = 0.192; 1GOX: δS = 0.288–0.691, S(a) = 0.288, S(b) = 0.290, S(c) = 0.294, S(d) = 0.307, S(e) = 0.311, S(f) = 0.315; 2TS1: δS = 0.227–0.687, S(a) = 0.256, S(b) = 0.273, S(c) = 0.284; 3GRS: δS = 0.154–0.688, S = 0.245; 451C: δS = 0.142–0.688, S = 0.237; 2CCY A: δS = 0.094–0.672, S(a) = 0.094, S(b) = 0.178; 2CRO: δS = 0.259–0.637, S = 0.259; 4MDH: δS = 0.166–0.690, S = 0.261; 8ADH: δS = 0.153–0.690, S = 0.292; 7RSA: δS = 0.326–0.689, S(a) = 0.350, S(b) = 0.508, S(c) = 0.533; 1HFX: δS = 0.341–0.691, S(a) = 0.435, S(b) = 0.490, S(c) = 0.520; 2CI2: δS = 0.300–0.684, S = 0.428; 2MM1: δS = 0.264–0.686, S(a) = 0.264, S(b) = 0.378, S(c) = 0.451, S(d) = 0.460, S(e) = 0.466, S(f) = 0.524, S(g) = 0.616, S(h) = 0.616; BARN: δS = 0.261–0.689, S(a) = 0.376, S(b) = 0.558; 132L: δS = 0.190–0.688, S(a) = 0.301, S(b) = 0.494, S(c) = 0.556, S(d) = 0.647; 1HRC: δS = 0.244–0.688, S(a) = 0.296, S(b) = 0.374, S(c) = 0.586, S(d) = 0.644. The helices marked a correspond to helical structures with the lowest entropy. They coincide with or are close to the absolute minimum of the entropy profile (see text). We have included as many segments as needed to scan all the α-helices assigned to each protein in the literature. Our numeration for 2CI2 is shifted ahead by 19 residues with respect to the one in ref. 60.
A: δS = 0.094–0.672, S(a) = 0.094, S(b) = 0.178; 2CRO: δS = 0.259–0.637, S = 0.259; 4MDH: δS = 0.166–0.690, S = 0.261; 8ADH: δS = 0.153–0.690, S = 0.292; 7RSA: δS = 0.326–0.689, S(a) = 0.350, S(b) = 0.508, S(c) = 0.533; 1HFX: δS = 0.341–0.691, S(a) = 0.435, S(b) = 0.490, S(c) = 0.520; 2CI2: δS = 0.300–0.684, S = 0.428; 2MM1: δS = 0.264–0.686, S(a) = 0.264, S(b) = 0.378, S(c) = 0.451, S(d) = 0.460, S(e) = 0.466, S(f) = 0.524, S(g) = 0.616, S(h) = 0.616; BARN: δS = 0.261–0.689, S(a) = 0.376, S(b) = 0.558; 132L: δS = 0.190–0.688, S(a) = 0.301, S(b) = 0.494, S(c) = 0.556, S(d) = 0.647; 1HRC: δS = 0.244–0.688, S(a) = 0.296, S(b) = 0.374, S(c) = 0.586, S(d) = 0.644. The helices marked a correspond to helical structures with the lowest entropy. They coincide with or are close to the absolute minimum of the entropy profile (see text). We have included as many segments as needed to scan all the α-helices assigned to each protein in the literature. Our numeration for 2CI2 is shifted ahead by 19 residues with respect to the one in ref. 60.