Figure 1.
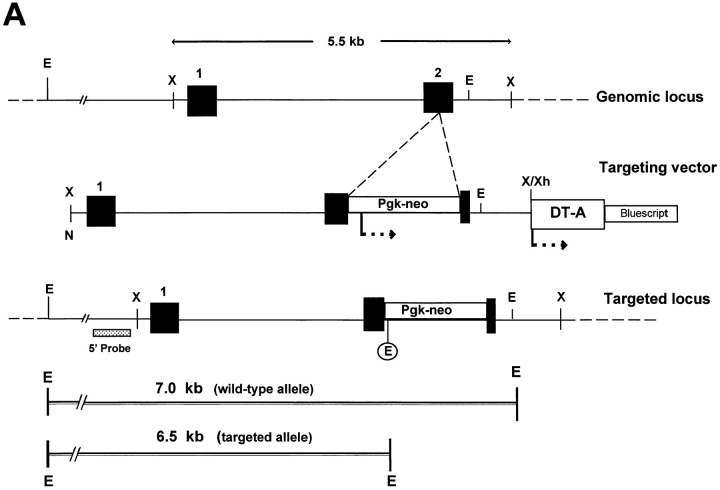
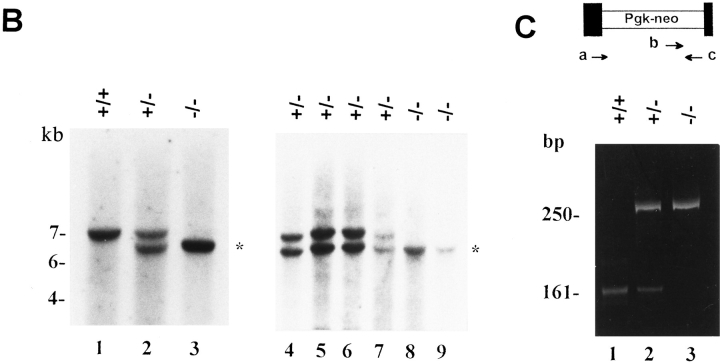
Disruption of the decorin gene locus in mouse ES cells and generation of decorin-deficient mice. (A) Targeting strategy. A 5.5-kb XbaI genomic fragment encoding exons 1 and 2 (filled boxes, not in scale) was used to construct the targeting vector. The predicted structure of the disrupted allele (bottom panel) shows the 5′ probe used to detect the diagnostic allele of 6.5 kb in contrast with the wild-type allele of 7.0 kb. This was due to the presence of a new EcoRI site in the Pgk-neo cassette. Abbreviations for restriction endonucleases: E, EcoRI; X, XbaI; N, NotI; Xh, XhoI. (B) Southern blot analysis of tail DNA isolated from two separate litters of mice including wild-type (+/ +), heterozygous (+/−), and homozygous (−/−) animals. Lanes 1–3 are from 3-mo-old animals derived from the breeding of two heterozygous mice, while lanes 4–9 are from newborn animals derived from the breeding of a heterozygous male and a homozygous female. The targeted allele of 6.5 kb is labeled by an asterisk. The DNA was separated on a 0.75% agarose gel, transferred to a nitrocellulose filter, and hybridized under high stringency to a PCRgenerated probe 5′ to the targeting vector. The size of molecular weight markers is indicated in the left margin in kb. (C) PCR detection of the targeted allele using primers specific for exon 2 (a and c, top scheme) or Pgk-neo (primer b). By using primer a and c, a fragment of 161 bp was identified in the wild-type (lane 1) and heterozygous animal (lane 2). However, the combination of primer b and c gave rise to a larger fragment of 250 bp, encompassing the 3′ end of the neo cassette and the 3′ end of exon 2, which was detected only in the heterozygous (lane 2) and homozygous (lane 3) animals. The products were separated on a 6% nonreducing polyacrylamide gel and stained with ethidium bromide. The size in bp is indicated in the left margin.