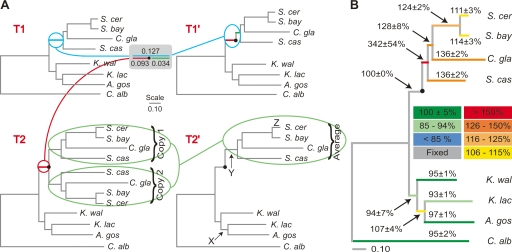
Figure 1.
Measuring the increase in the rate of protein sequence evolution after gene duplication. (A) Construction of a pair of topologically identical trees, T1′ and T2′ (right), from a tree derived from single-copy sequences only (T1; obtained from super-alignment A1′) and a tree derived from single- and double-copy sequences (T2; obtained from super-alignment A2′). The tree T1′ was derived from the tree T1 by partitioning the branch between the divergence of the non-WGD yeasts and the divergence of S. castellii from the S. cerevisiae lineage into pre- and post-duplication segments (light-blue line and gray box). As in Scannell et al. (2006), we assumed that the length of the preduplication branch on T1 is the same as that on T2 (red line). The tree T2′ was derived from the tree T2 by averaging the lengths of all duplicated branches between the post-WGD clades labeled “Copy 1” and “Copy 2” (light-green ovals). A filled circle (●) indicates the inferred point of duplicate gene divergence. The branches labeled X, Y, and Z on T2′ are referred to in the text. (B) Tree showing the length of branches on T2′ as a percentage of the length of the corresponding branches on T1′, which is a measure of the rate of evolution of double-copy sequences relative to single-copy sequences. Percentages (± one standard deviation) are averages from 100 bootstrap replicates (see Methods). The branch lengths drawn are the averages on T1′ from the same 100 bootstrap replicates. Branches are colored according to the arbitrary scale shown.