Abstract
The Bacillus subtilis YxiN protein is a modular three-domain RNA helicase of the DEx(D/H)-box protein family. The first two domains form the highly conserved helicase core, and the third domain confers RNA target binding specificity. Small angle x-ray scattering on YxiN and two-domain fragments thereof shows that the protein has a distended structure in solution, in contrast to helicases involved in replication processes. These data are consistent with a chaperone activity in which the carboxy-terminal domain of YxiN tethers the protein to the vicinity of its targets and the helicase core is free to transiently interact with RNA duplexes, possibly to melt out misfolded elements of secondary structure.
The DEx(D/H)-box RNA helicases are modular, multidomain proteins that effect a diversity of structural rearrangements of RNA (1). Proteins of the DEAD-box protein A (DbpA) subfamily of the DEx(D/H)-box helicases facilitate ribosome maturation in a manner that is not yet well understood. DbpA homologs consist of a conserved catalytic fragment, the N-domain of which binds ATP, plus a small, 76-residue carboxy-terminal RNA binding domain (RBD) which is unique to, and defines, the DbpA subfamily (2) (Fig. 1). A question arises whether the domains of DbpA proteins normally form a stable, compact structure in solution, similar to replication-involved helicases such as Rep (3), PcrA (4), and the hepatitis C virus helicase fragment (5), or whether they are distended from each other like beads on a string, and only condense into a (stable or transient) compact structure in the presence of the ligands MgATP and/or RNA. Although posed as a structural question, this bears direct relevance to the mechanism and activity of DbpAs (discussed below). A precedent for a DEx(D/H)-box helicase in a distended conformation is provided by the crystal structure of yeast eIF4A (6), where the two helicase domains are tethered loosely to each other by an extended peptide linker, with no intramolecular interaction between the domains. In this context, we have used solution small-angle x-ray scattering to examine the solution conformations of a DbpA protein, the three-domain YxiN protein of Bacillus subtilis (2) and subfragments thereof.
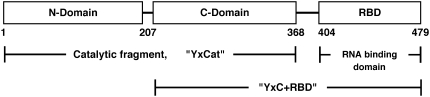
FIGURE 1.
Schematic drawing of the three-domain structure of the B. subtilis YxiN protein and subfragments used in these experiments, with amino-acid residue numbers of boundaries of recombinant expression constructs.
A schematic drawing of the primary structure of YxiN, along with a delineation of the nomenclature and protein fragments used in these experiments, is shown in Fig. 1. Measurements were made on full-length YxiN and the two-domain catalytic fragment YxCat in the presence of MgATP, for proteins in which Glu152, the presumed catalytic residue of ATP hydrolysis, was mutated to glutamine (E152Q) to prevent nucleotide hydrolysis. Measurements were also made on wild-type YxiN. Aggregation of YxiN and YxCat at high protein concentrations in the presence of MgADP or absence of nucleotide precluded measurement of scattering data under those conditions. Experiments were also carried out on the two-domain fragment YxC+RBD, which lacks the ATP-binding N-domain; nucleotide was not included in the samples. Results are summarized in Table 1, and experimental details are provided in the Supplementary Material.
TABLE 1.
Derived solution scattering parameters for YxiN and subfragment
| Beamline/protein | Concentrations* (mg/ml) | Nucleotide | Resolution† (Å) | dmax (Å) | Rg (Å) |
|---|---|---|---|---|---|
| APS BL 18-ID | |||||
| YxiN(E152Q) | 1.25; 2.5; 5.0 | 10 mM MgATP by addition | 66.7 | 130 ± 5 | 35.7 ± 0.2 |
| YxiN(E152Q) | 2.5; 5.0 | 10 mM MgATP by dialysis | 35.7 | 124 ± 4 | 34.7 ± 0.1 |
| YxiN(E152Q) | 1.25; 2.5; 5.0 | 1 mM MgATP by dialysis | 14.1 | 130 ± 5 | 33.6 ± 0.1 |
| YxCat(E152Q) | 1.4; 2.8; 5.6 | 10 mM MgATP by dialysis | 16.5 | 95 ± 5 | 29.0 ± 0.2 |
| YxC+RBD | 2.5; 10.0 | None | 13.4 | 95 ± 5 | 26.3 ± 0.2 |
| SSRL BL 4-2 | |||||
| YxiN | 2.5; 5.0; 10.0 | 1 mM MgATP by dialysis | 24.2 | 130 ± 5 | 37.3 ± 0.5 |
| YxC+RBD | 10 | None | 59.5 | 95 ± 5 | 25.3 ± 0.7 |
Includes the concentrations of all protein samples whose scattering curves were merged together as described in Supplementary Material.
Resolution = λ/2sin(θ), where λ is the wavelength and 2θ is the largest scattering angle for which data were used.
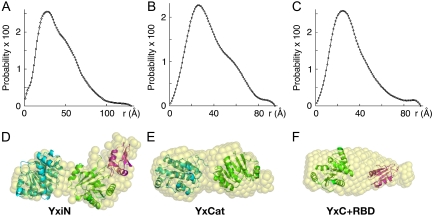
Pair-distance distribution functions (i.e., P(r) plots, Fig. 2) were computed from the scattering curves (Fig. S1, Supplementary Material). For YxiN and YxCat, the P(r) curves show a clear secondary bump at increasing distance after the peak, a feature consistent with a solution structure in which two or more domains are distended from each other rather than condensed into a compact assembly. Low-resolution molecular shapes, in the form of assemblies of dummy atoms, were computed and averaged using the programs DAMMIN and DAMAVER (7,8) as described in the Supplementary Material's Methods, providing envelopes for the average scattering density. χ-values, measuring the goodness of fit between the scattering curve computed from the assembly of dummy atoms and the experimental scattering curve, were 1.75, 0.88, and 1.28 for YxiN, YxCat, and YxC+RBD, respectively. The molecular envelopes for all three constructs are multilobed.
FIGURE 2.
Distance distribution (P(r)) plots (A–C) and solution models (D–F) for YxiN (A and D), YxCat (B and E), and YxC+RBD (C and F). Distance distribution plots have 100 points and are normalized such that the sum of all probability values is unity. Pseudo atoms of the molecular shapes (D–F) computed with the programs DAMMIN (7) and DAMAVER (8) are represented as semitransparent spheres. Results of rigid body optimization of the relative positions of crystallographic models of domains using the program SASREF (9) shown in panels D–F as ribbon drawings with color coding: N-domain of eIF4A, cyan; YxiN C-domain, green; YxiN RBD, magenta. The multidomain fragments from SASREF have been manually placed within the molecular envelopes of the molecular shapes to visualize agreement between the two results. Panels D–F made with the program PYMOL (W. L. Delano, “The Pymol Molecular Graphics System”, http://pymol.sourceforge.net).
Crystallographic structures of individual domains (YxiN RBD, PDB 2G0C; YxiN C-domain, PDB 2HJV; since the N-domain of YxiN is unknown, the N-domain of eIF4A, which is 38% identical in sequence, PDB 1QVA) were used in computational rigid body refinement of their optimal relative orientation and separation consistent with the scattering data using the program SASREF (9). In each case, the best fit of relative dispositions of the domains making up each fragment place the domains at significant distances from each other. For this model of YxCat, the computed centroid-to-centroid distance between the two domains is 45.4 Å; in the crystal structure of eIF4A, it is 48.6 Å. This single-conformation model of YxCat in solution is distended, similar to eIF4A in crystals.
Similar distended conformations result for YxiN and the two-domain YxC+RBD fragment, although the features in the P(r) plot and molecular envelope are less pronounced in the latter case. Manual placement of the models from SASREF within the molecular envelopes from DAMMIN shows general agreement between the results from the independent computational methods (Fig. 2). Nonetheless, some of the molecular envelopes, particularly for YxC+RBD, appear to have extra density not filled by the models.
To accommodate multiple conformations in the modeling, an ensemble optimization method was used in which ensembles of models with linkers of variable (flexible) conformations between domains are selected which optimize agreement between the computed composite ensemble scattering and experimental data (10). These calculations resulted in ensembles of structures whose mean Rg and dmax values agree with those from the other analyses, but which have a distribution of conformations (Fig. S2). For the two-domain constructs YxCat and YxC+RBD, the ensemble of structures generally fits the molecular envelope that results from model-independent analysis of the data and account for the extra envelope density that is not well fit with a single model (Fig. S2, K and L). The apparent fit is less good for YxiN, as expected in view of the extended linker (35 residues) and low scattering contribution (16%) of the RBD. These results corroborate a model in which the domains of YxiN may contact each other transiently, but are generally distended in solution.
A paradigm for the structure and mechanism of processive replication helicases is that compact, multidomain molecules thread a single strand oligonucleotide between domains; one attractive hypothesis is that they work through an inchworm mechanism in which the protein translocates along one strand of DNA or RNA in an ATP-driven reaction, displacing the second strand in the process (11). In contrast, despite sharing a sequence and structural similarity with replication helicases in the catalytic fragment, many DEx(D/H)-box RNA helicases are nonprocessive (12,13) and appear to function differently. The crystallographic structure of the Vasa DEAD-box helicase catalytic fragment with U10 bound (14) suggests that the protein functions by binding and kinking one strand of RNA on one side of the catalytic fragment; such an interaction could destabilize an RNA duplex. This is consistent with this group of helicases working as RNA chaperones that facilitate melting of short segments of unstable RNA duplexes through a transient, nonprocessive duplex destabilization activity.
The data presented here, demonstrating that the domains of YxiN are loosely tethered and distended in absence of ligands, support a model in which YxiN functions as an RNA chaperone. The RBD would target YxiN to the nascent catalytic site of the 50S subunit of the ribosome by binding tightly to hairpin H92 of 23S rRNA. Then, the catalytic fragment could search locally for segments of unstable, misfolded RNA and transiently disrupt them, allowing rapid and repetitive cycles of unfolding and refolding until the correct structure is achieved.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL) and the Advanced Photon Source (APS). We thank Dr. Hiro Tsuruta (SSRL beamline (BL) 4-2), Dr. Sonke Seifert (APS BL 12-ID-C), and Liang Guo (APS BL-18-ID) for valuable staff support.
Supported by National Institutes of Health grant GM-71696 (D.B.M.) and a fellowship from the Alfred Benzon Foundation (M.T.O.).
Michael T. Overgaard's current address is Department of Molecular Biology, University of Aarhus, Denmark.
Editor: Kathleen B. Hall.
References
- 1.Rocak, S., and P. Linder. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5:232–241. [DOI] [PubMed] [Google Scholar]
- 2.Kossen, K., and O. C. Uhlenbeck. 1999. Cloning and biochemical characterization of Bacillus subtilis YxiN, a DEAD protein specifically activated by 23S rRNA: delineation of a novel sub-family of bacterial DEAD proteins. Nucleic Acids Res. 27:3811–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korolev, S., J. Hsieh, G. H. Gauss, T. M. Lohman, and G. Waksman. 1997. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 90:635–647. [DOI] [PubMed] [Google Scholar]
- 4.Subramanya, H. S., L. E. Bird, J. A. Brannigan, and D. B. Wigley. 1996. Crystal structure of a DExx box DNA helicase. Nature. 384:379–383. [DOI] [PubMed] [Google Scholar]
- 5.Yao, N., T. Hesson, M. Cable, Z. Hong, A. D. Kwong, H. V. Le, and P. C. Weber. 1997. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 4:463–467. [DOI] [PubMed] [Google Scholar]
- 6.Caruthers, J. M., E. R. Johnson, and D. B. McKay. 2000. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. USA. 97:13080–13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svergun, D. I. 1999. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76:2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volkov, V. V., and D. I. Svergun. 2003. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36:860–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petoukhov, M. V., and D. I. Svergun. 2005. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 89:1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernadâo, P., E. Mylonas, M. V. Petoukhov, M. Blackledge, and D. I. Svergun. 2007. Structural characterization of flexible proteins using small-angle x-ray scattering. J. Am. Chem. Soc. 129:5656–5664. [DOI] [PubMed] [Google Scholar]
- 11.Velankar, S. S., P. Soultanas, M. S. Dillingham, H. S. Subramanya, and D. B. Wigley. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 97:75–84. [DOI] [PubMed] [Google Scholar]
- 12.Rogers, G. W., Jr., N. J. Richter, and W. C. Merrick. 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274:12236–12244. [DOI] [PubMed] [Google Scholar]
- 13.Rogers, G. W., Jr., W. F. Lima, and W. C. Merrick. 2001. Further characterization of the helicase activity of eIF4A. Substrate specificity. J. Biol. Chem. 276:12598–12608. [DOI] [PubMed] [Google Scholar]
- 14.Sengoku, T., O. Nureki, A. Nakamura, S. Kobayashi, and S. Yokoyama. 2006. Structural basis for RNA unwinding by the DEAD-box protein Drosophila vasa. Cell. 125:287–300. [DOI] [PubMed] [Google Scholar]