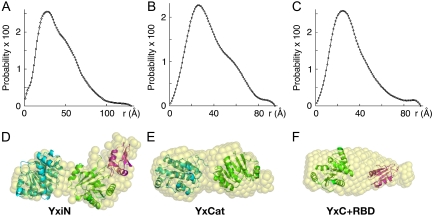
FIGURE 2.
Distance distribution (P(r)) plots (A–C) and solution models (D–F) for YxiN (A and D), YxCat (B and E), and YxC+RBD (C and F). Distance distribution plots have 100 points and are normalized such that the sum of all probability values is unity. Pseudo atoms of the molecular shapes (D–F) computed with the programs DAMMIN (7) and DAMAVER (8) are represented as semitransparent spheres. Results of rigid body optimization of the relative positions of crystallographic models of domains using the program SASREF (9) shown in panels D–F as ribbon drawings with color coding: N-domain of eIF4A, cyan; YxiN C-domain, green; YxiN RBD, magenta. The multidomain fragments from SASREF have been manually placed within the molecular envelopes of the molecular shapes to visualize agreement between the two results. Panels D–F made with the program PYMOL (W. L. Delano, “The Pymol Molecular Graphics System”, http://pymol.sourceforge.net).