Fig. (6).
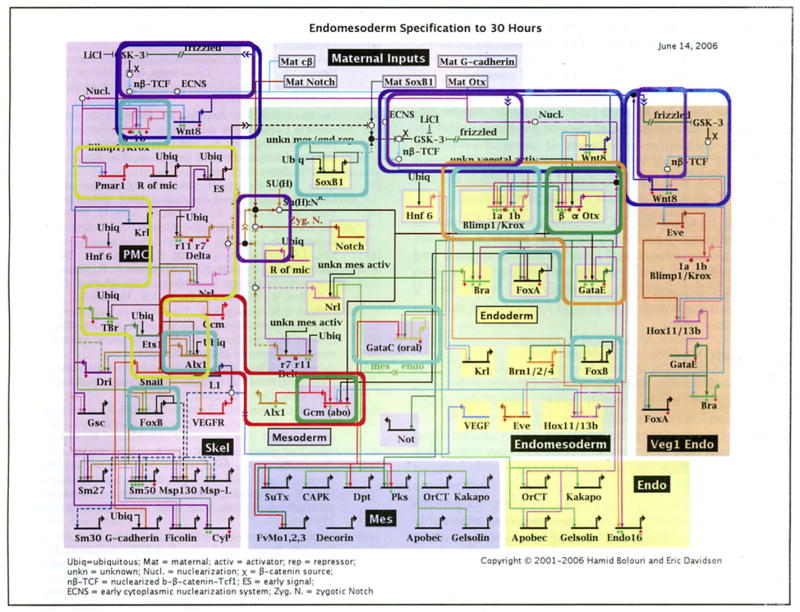
Occurrences of putative functional building blocks in the GRN underlying endomesoderm specification in the embryo of the sea urchin Strongylocentrotus purpuratus. Each occurrence is indicated by a rounded bounding box. Box colors identify different types of functional building blocks. Green: single-gene intra-cellular positive feedback latches. Orange: a multi-gene intra-cellular positive feedback latch. Dark blue: inter-cellular positive feedback latch (the community effect) mediated by Wnt8 signaling. Cyan: instances of negative auto-regulation. foxA expression is oscillatory (Oliver! P, Tu Q, Walton K, Davidson EH and McClay DR, manuscript in preparation). The expression patterns of the other auto-repressive genes are consistent with the ‘single pulse’ functional building block. Purple: signal mediated toggle switches mediated via β-catenin and TCF/LEF in Wnt signaling and Su(H) in Notch signaling. Red: the alx1-gcm mutual exclusion operator. Alx1 is on in the PMC domain and off in the mesoderm. Gcm is on in the mesoderm and off in the PMC. Yellow: the pmar1 gradient detection/analogue to digital switch. Pmar1 represses an uncharacterized repressor which in turn represses es, delta, nrs, alx1, tbr and ets1.
Each short horizontal line from which a bent arrow extends to indicate transcription represents the cis-regulatory element that is responsible for expression of the gene named in the domain shown. The arrows and barred lines indicate the normal function of the gene (activation or repression) as deduced from changes in transcript levels in perturbation experiments. Red diamonds indicate that a cis-regulatory element, which controls relevant expression has been isolated. Green diamonds indicate reported experimental evidence validates expected target site. The rectangles in the lower tier of the diagram show downstream differentiation genes. Dashed lines indicate inferred or possibly indirect relationships. Arrows inserted in arrow tails indicate intercellular signaling interactions. Large open ovals represent cytoplasmic biochemical interactions at the protein level. Sec [4] for a detailed description.
