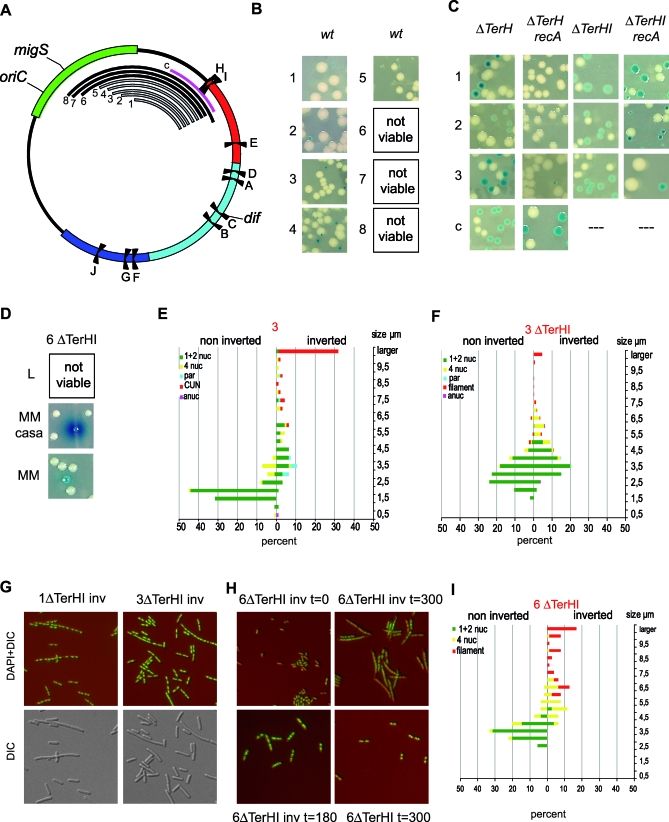
Figure 5. Intrareplichore Inversions between the Right and Ori MDs.
(A) Schematic representation of the E. coli chromosome showing MDs, Ter sites, oriC, migS, and dif. The numbered black arcs indicate the position and extent of fragments inverted in strains 1 to 8 and (1 to 8: intra O-R1 to −8 in Table 2), and the control strain c is indicated in pink (control Intra R-NSright3 in Table 1).
(B) Colonies of strains carrying chromosomes with various intrareplichore inversions as indicated in (B). Inversion tests for strains 1 to 8 were performed in rich medium. Colonies carrying the inverted configuration of strains 1 to 5 can be obtained in rich medium (indicated by a grey arc), whereas colonies carrying the inverted configuration of strains 6 to 8 cannot be obtained (indicated by a black arc).
(C) Colonies of strains 1 to 3, in the absence of TerH or TerH and TerI, carrying the chromosome in the wt or inverted configurations. Colonies were obtained in a wt or in a recA background. Also shown are the results obtained after deletion of the TerH site from strain c in a wt or recA genetic background.
(D) Colonies of strain 6 deleted for TerH and TerI, in the wt or inverted configuration, obtained in minimal medium (MM) or minimal medium supplemented with casamino-acids (MM casa).
(E) Microscopic analysis of strain 3 carrying a chromosome in the wt (noninverted) and inverted configurations as described in Figure 2.
(F) Nucleoid and cell analysis obtained from strain 3 deleted for TerH and TerI, carrying a chromosome in the wt or inverted configuration. The left and right colored horizontal bars indicate the percentage of the different types of cells and nucleoids in the wt and inverted configurations, respectively (green indicates cells containing one and two nucleoids; yellow: cells containing four nucleoids; cyan: cells containing par-like nucleoids; red: filamentous cells with apparently segregated nucleoids; and pink: anucleate cells).
(G) Images of combined DIC-DAPI and DIC-stained cells from strains 1 and 3 deleted for TerH and TerI, in the inverted configurations, showing the presence of filamentous cells with segregated nucleoids.
(H) Combined images of DIC and DAPI pictures of cells from strain 6 deleted for TerH and TerI, in the inverted (inv) or wt configuration after growth in rich medium for 0, 180, or 300 min.
(I) Nucleoid and cell analysis obtained from strain 3 deleted for TerH and TerI, carrying a chromosome in the wt or inverted configuration, after 300 min of growth in rich medium. The left and right colored horizontal bars indicate the percentage of the different types of cells and nucleoids in the wt and inverted configurations, respectively (green indicates cells containing one and two nucleoids; yellow: cells containing four nucleoids; and red: filamentous cells with un-segregated nucleoids).