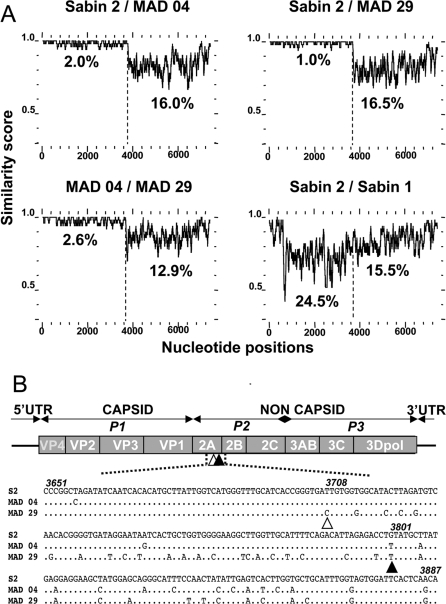
Figure 2. Genomic Features of the Poliovirus Isolates.
(A) Plots of similarity between a set of aligned genomes (Sabin 2, MAD 04, MAD 29, and Sabin 1), using a 50-nt sliding window. Approximate nucleotide positions in the poliovirus genome are indicated. Average nucleotide identities between the 5′ vaccine part and the 3′ non-vaccine part of the recombinant genomes are shown.
(B) Sites of the recombination junctions in the recombinant VDPV genomes. The genetic organization of the PV genome is shown, including the 5′- and 3′-UTR. Genomic regions P1 to P3 encoding viral proteins (VP4 to 3Dpol) are indicated. Open and closed triangles indicate the approximate sites of the recombination junctions of the MAD 29 and MAD 04 recombinant genomes, respectively. Both junctions are located at the 3′ end of the protease 2A genomic region. MAD 04 and MAD 29 sequences (nt 3651–3890) on both sides of the recombination junctions are given and compared with Sabin 2 sequences (S2). Nucleotide numbering according to Sabin 2 sequences is given in italics.