Abstract
Evidence indicates that oxidative stress inhibits cell proliferation in several cell systems. To determine whether the proliferation of Caco-2 cells is inhibited by oxidative stress and to identify any novel key regulatory factors involved in protecting or damaging the intestine from oxidative stress, Caco-2 cells were treated with an oxidizing agent and analyzed by transcriptomic oligonucleotide microarrays. Results indicated that expression of genes involved in cell proliferation and growth, including genes involved in lipid synthesis, cell cycle progression and cell division, angiogenesis, RNA processing and translation, cAMP metabolism, cytoskeleton and cell to cell adhesion, receptor tyrosine kinases, and intracellular and extracellular signaling, were repressed. If an oxidant-induced inhibition in cell proliferation is involved in the pathogenesis of intestinal disease, information gained could help explain the mechanisms contributing to the causes and consequences of intestinal disease and could aid in the elucidation of mechanisms by which intestinal cells protect against oxidative stress.
Keywords: Caco-2 cells, Oxidation, Cell proliferation, Microarray
Introduction
Reactive oxygen species (ROS), such as hydrogen peroxide (H2O2), are implicated in the origination and development of inflammatory bowel disease (IBD), ischemia-reperfusion injury, intestinal cancer, and other chronic intestinal disorders [1, 2]. The intestinal epithelium is continuously exposed to ROS from xenobiotics, toxins, catalase-negative bacteria, cast-off mucosal cells, and ingested food [1]; therefore, factors that protect intestinal epithelial cells from oxidative stress and that are responsible for the manifestation of the epithelium’s integrity must be identified. Over evolutionary time, respiring organisms and specific tissues in these organisms were forced to develop protective mechanisms that could maintain the level of oxidation or damage from oxygen-derived free radicals, such as superoxide anion, H2O2, and the hydroxyl radical low. The gastrointestinal (GI) tract is equipped with defense systems designed to eliminate potentially damaging and disease causing ROS, H2O2; although the exact mechanism is not completely understood.
The human colonic adenocarcinoma cell line, Caco-2 cells, is widely used in research as an in vitro intestinal model [2]. Caco-2 cells differentiate spontaneously when grown to confluence [2, 3]. This differentiation is a function of time. Even though this cell line is derived from colon cancer cells, Caco-2 cells begin resembling normal small intestinal epithelial cells or enterocytes at about 4 days after confluence [4]. Although some differences exist between Caco-2 cells and human tissue [5], other research [3] has demonstrated the use of Caco-2 cells as a feasible model for in vivo intestinal studies. For example, the gene expression of certain enzymes in Caco-2 cells was similar to the gene expression of these same enzymes in human intestinal resections. More-over, transcriptome changes during differentiation of Caco-2 cells were comparable to the changes found in mouse small intestinal crypt and villous cells [3].
Oxidative stress occurs in a cell or organism when there is an imbalance in the generation and removal of ROS [6]. Hydrogen peroxide can damage DNA, lipids, and proteins if there is a concentration of more ROS than the organism’s antioxidant systems can dispose of efficiently [6, 7]. Many studies have exemplified the role of H2O2 in the generation of destructive free radicals and DNA damage [2, 7, 8]. To keep destructive free radical levels low, H2O2 can be metabolized by the intrinsic antioxidant enzymes, catalase and glutathione peroxidase (GPX) [8], in the intestine, but other defense mechanisms for the intestine exist. Evidence indicates that oxidative stress induces inhibition of cell proliferation in a number of cell systems [1]. This oxidant-induced inhibition of cell proliferation may be involved in the pathogenesis of intestinal disease [1]. Therefore, prevention of damage by oxidative stress or H2O2 must also involve cell rescue and defense processes that help maintain cellular integrity by proper equilibrium between DNA repair, DNA replication, cell proliferation, apoptosis, stress response, and other conditions that allow the cell to prevent and cope with oxidation [1, 6].
To identify any novel key regulatory factors involved in protecting or damaging the intestine from oxidative stress, H2O2 was used to produce oxidative stress in Caco-2 cells followed by a transcriptomic analysis using microarrays. Transcription of genomic DNA to produce mRNA is the first step in the process of protein synthesis and is indicative of a cell’s response to some environmental stimulus, in this case H2O2. Other microarray studies [6, 9-13] have successfully provided information about cellular processes and molecular mechanisms of organisms. To seek links between gene expression patterns and the processes of intestinal disease development, this study analyzed the change in expression of approximately 13,000 human genes in Caco-2 cells after treatment with H2O2 through the use of an oligonucleotide microarray. It is expected that the collected data will improve the understanding of the consequences of oxidation to the intestine. This information may also be important in preventing intestinal disease and improving health, perhaps by the manipulation of specific molecular targets with selective nutritional antioxidant agents in individuals at risk for intestinal disease.
Materials and methods
Cell culture
Caco-2 cells (ATCC, Manassas, VA) were maintained by serial passage in 75-cm2 culture flasks at 37°C and 85-90% relative humidity in an incubator supplied with 5% CO2.The cells were grown in 15 ml of Dulbecco’s Modified Eagle Medium (DMEM; GIBCO™, Grand Island, NY) containing, per volume, 20% fetal bovine serum, 1% l-glutamine, 1% penicillin/streptomycin, and 1% nonessential amino acids. New culture medium was supplied every 48 hr. After attaining confluence, the cells were harvested using trypsin and split (1:2).
Flow cytometry
Flow cytometry was used to determine the appropriate concentration of H2O2 to use as the oxidizing treatment on the Caco-2 cells without producing cytotoxic effects. The concentration of the selected H2O2 level corresponded to > 95% cell viability. Cells were grown until confluent. Duplicate samples of control and 50 μM, 75 μM, and 100 μM H2O2-treated cells were incubated at 37°C for 30 min and were collected after trypsinization in DMEM. Total number of cells per millimeter in control flasks was calculated using a hemocytometer. After pelleting by centrifugation, cells were fixed with 70% ethanol and resuspended in 0.1% Triton-X solution to obtain 1 million cells/ml. To 2 ml of cells, 5 μg/ml propidium iodide and 5 μg/ml RNase for DNA staining were added. The cells were placed on ice until analysis [14]. Stained cells were analyzed by fluorescence-activated cell scan (FACS; Becton Dickinson, San Jose, CA) flow cytometry and ModFit software to determine the number of viable cells, cell cycle phase distribution, and percentage of nuclei with hypodiploid content, which is indicative of apoptosis-associated nonrandom DNA fragmentation [14].
H2O2 treatment of cells
A 100 mM stock solution of H2O2 (Sigma Chemical, St. Louis, MO) was diluted to a final concentration of 75 μM serum-free DMEM (SFDMEM) containing, per volume, 1% l-glutamine, 1% penicillin/streptomycin, and 1% nonessential amino acids. Medium was aspirated from 16 flasks of Caco-2 cells (passage 44) and cells were washed with phosphate-buffered saline (PBS). Fifteen milliliters of supplemented SFDMEM was added to eight control flasks, which were then incubated at 37°C with 5% CO2 for 30 min. Fifteen milliliters of 75 μM H2O2 in supplemented SFDMEM was added to eight treatment flasks, which were then incubated at 37°C with 5% CO2 for 30 min. After incubation, medium was aspirated and cells were washed with PBS.
RNA isolation and purification
Total RNA was isolated from each sample using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions [5, 11]. RNA from eight control flasks was pooled, as was RNA from eight treatment flasks [10, 15, 16]. The total RNA was further purified using a Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA) [5, 11, 12]. The quality and quantity of the RNA samples were assessed by measuring the optical density of each sample at 260 nm followed by 280 nm (UV Mini 1240 Spectrophotometer; Shimadzu, Kyoto, Japan) [13]. To confirm the integrity of RNA, 10 μg of RNA from each sample was electrophoresed through a 1% agarose/formaldehyde gel containing ethidium bromide for approximately 2 hr. The 18S and 28S RNA bands were observed and no RNA degradation was detected [13]. The RNA samples were stored at -70°C for microarray and reverse transcription-polymerase chain reaction (RT-PCR) analysis.
Microarray analysis
All procedures regarding the microarray completion and analysis were performed at the Genomics Core Facility at the Center for Biotechnology at the University of Nebraska—Lincoln. Using the purified total RNA from the control and H2O2 samples as the templates, double-stranded complementary DNA (ds cDNA) was synthesized according to previously described protocols [5]. From the ds cDNA, biotinlabeled cRNA was synthesized and prepared for microarray hybridization [5]. Samples were then hybridized onto separate Human U133A GeneChips® (Affymetrix, Santa Clara, CA) according to the Affymetrix protocol. Using the GeneChip® Fluidics Station 450, the arrays were washed, then stained with streptavidin-phycoerythrin (SAPE), and the signals were amplified by staining with a biotinylated, anti-streptavidin antibody solution [5]. Fluorescence was measured with the Affymetrix GeneChip® Scanner 3000. Gene expression profiles were analyzed using the Affymetrix Microarray Suite (MAS) and Data Mining Tool (DMT) software. This GeneChip® software by Affymetrix processed array images from the scanner, corrected for background variation, normalized arrays for array-to-array comparison, and calculated statistics to indicate whether a gene transcript was present, marginally present, or absent, and to indicate whether a treatment gene transcript was significantly differentially expressed from the control [5, 11, 12].
Because a large range of gene intensities was exhibited, the data were transformed to a log scale [10]. Results are expressed as a change, either increase or decrease, in expression level relative to the control for each gene. To minimize sample-to-sample variability, due to cell culture, extraction and purification of mRNA, cDNA synthesis, cRNA labeling, hybridization procedures, and signal detection, and to verify results, two independent experiments were completed [11, 15]. The absolute detection call (present, marginal, absent) and detection p-value were determined for each probe set in control and H2O2 treatment arrays. A probe set, which represents a particular gene, was eliminated from analysis when the absolute detection call was absent on both control and H2O2 treatment arrays in either or both of the experiments. The relative change call (increase, decrease, or no change), change p-value, and signal log ratio between H2O2-treated and control samples were then determined for each probe set. It was obligatory for probe sets to have a relative consistent change of intensity (increased or decreased) in both experiments. The average difference of gene expressions between control and H2O2-treated samples or signal log ratio of the two experiments was then calculated using DMT. Finally, all probe sets showing a change of expression in both experiments and having an average signal log ratio or fold change ≥0.40 or ≥1.32, respectively, were identified [10]. Genes represented by these probe sets were identified as the most consistently detected and most significantly changed after H2O2 treatment.
RT-PCR
Confirmation of the expression changes observed in the microarrays of genes of interest was analyzed by semi-quantitative RT-PCR [5, 11, 12]. Histone 1, H4F (H4FC), a gene that had no obvious change in mRNA levels during the experiment, was chosen for normalization in the RT-PCR [11]. From 1 μg of control and H2O2 treatment total RNA, synthesis of cDNA via reverse transcription was accomplished using SuperScript™ III (Invitrogen, Carlsbad, CA) [11]. The reverse-transcribed generated cDNA was amplified by PCR using gene-specific primers of genes of interest as presented in Table 1. One-microliter aliquots of the control and H2O2 treatment cDNA were amplified separately with the specific forward and reverse primers of the gene of interest and the BD Advantage cDNA PCR Kit (BD Biosciences Clontech, Palo Alto, CA) containing Polymerase Mix. For amplification, PCR was carried out as follows: 1 min at 95°C for one cycle and then denaturation for 1 min at 94°C, annealing for 1 min at 55°C, and elongation for 2 min at 72°C, repeated for 18 to 32 cycles depending on the gene of interest. This procedure was completed in order to obtain a linear correlation between the amount of PCR product and template cDNAs [5]. Products were electrophoresed through 1% agarose gels with ethidium bromide [12] and band intensities were quantified using Kodak1D software. Results of RT-PCR were compared to array data to evaluate if fold changes of gene expression between the two methods were comparable.
Table 1.
Primer sequences for RT-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| GSR: Glutathione reductase | AAGGGTCATATCATCGTAG | GTCTCCTGGTTCTCAACGA |
| IGF2: Insulin-like growth factor 2 | GGCAAGTTCTTCCAATATG | GGAGGGTATGTGAAGGGTG |
| FADS2: Fatty acid desaturase 2 | CTGAAATACCTGCCCTACAA | ATCTTGTGTAAGTTGTGCCG |
| GPX2: Gastrointestinal glutathione peroxidase | AGTCTCAAGTATGTCCGTC | CCTTTATTGGTCTCTTCTA |
| GPX4: Phospholipid hydroperoxidase | GAGGCAAGACCGAAGTAAAC | TTTATTCCCACAAGGTAGCC |
| CYP2B6: Cytochrome P450 2B6 | AAAGAGAAATCCAACGCACAC | ATCTGGTATGTTGGGGGTATT |
| H4FC: Histone 1, H4F | ATGTCTGGTAGAGGCAAAGGTGGTAAA | CGCACTCTGTACGGCTTTGGTGGCTGA |
Results
Flow cytometry
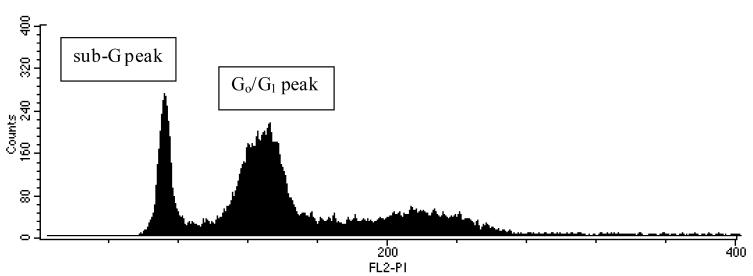
After treatment of the cells with increasing concentrations of H2O2 (50-100 μM), viable Caco-2 cells were compared with apoptosis-associated nonrandom DNA fragmentation by FACS flow cytometry. More than 11,000 modeled events were counted for each sample. As depicted in Fig. 1, the region designated the sub-G peak is indicative of apoptosis-associated nonrandom DNA fragmentation of cells treated with 50 μM H2O2. From these studies, it was determined that H2O2 induced these apoptotic events in Caco-2 cells in a concentration-dependent manner (data not shown)
Fig. 1.
Flow cytometry. The sub-G peak represents Caco-2 cells undergoing apoptosis-associated nonrandom DNA fragmentation after treatment with 50 μM H2O2
The average numbers of apoptotic events were 3.1%, 3.1%, and 4.5% in Caco-2 cells subjected to 50, 75, and 100 μM H2O2, respectively. Although all three H2O2 levels produced apoptotic events with averages less than the designated 5%, the 75 μM concentration was selected for two reasons. First, DNA damage was produced without significantly altering the viability of the cells. Second, one of the 100 μM concentration samples did exceed the 5% designated limit for apoptotic events. This was in agreement with Aherne and O’Brien [7], who found that the 75 μM H2O2 concentration did not significantly alter the viability of the cells but did produce significant DNA damage.
Microarray analysis
This study was conducted to evaluate the effects of H2O2 on the expression of 12,931 genes in Caco-2 cells. For the vast majority of transcripts, expression was unchanged after H2O2 treatment. From the two independent experiments, 108 probe sets, representing 87 genes of known identity, showed a change of expression. This represents < 1% of the total 12,931 genes represented on the array. There were 10 known genes detected that showed a change in expression, with redundant probe sets, and 7 unknown genes detected with a change in expression. Comparison of the expression patterns from the control and H2O2 groups showed repression of 85 genes and the induction of two genes for the treated cells. Genes that altered expression were categorized using Batch Query NetAffx (Affymetrix, Santa Clara, CA) according to their gene title, gene symbol, biological process, molecular function, cellular component, and pathway. A list of these 87 genes, their average signal log ratio change, and their corresponding biological process are listed in Table 2. Several genes involved in lipid synthesis, cell cycle and cell growth, angiogenesis, transcription and DNA repair, RNA processing and translation, protein processing and proteolysis, glucose production, cytoskeleton and cell-to-cell adhesion (Fig. 2), and cell signaling were repressed by H2O2.
Table 2.
List of affected genes
| Gene name | Average signal log ratio change | Biological process(es) |
|---|---|---|
| ADP-ribosylation factor 3 | -0.42 | Intracellular protein transport/small GTPase-mediated signal transduction |
| Apoptosis inhibitor 5 | -0.58 | Transport/anti-apoptosis |
| Aspartyl-tRNA synthetase | -0.46 | Protein biosynthesis |
| Ataxin 2-like | -1.12 | Neurodegenerative disorders |
| ATPase, H+ transporting, lysosomal | -0.69 | Hydrogen ion transporter activity/important for protein sorting |
| BCL2-associated X protein | -0.57 | Apoptosis |
| Carboxypeptidase D | -0.49 | Proteolysis and peptidolysis |
| CCR4-NOT transcription complex, subunit 3 | -0.59 | Regulation of transcription, DNA-dependent |
| Chorionic gonadotropin, β polypeptide 7 | -0.47 | Apoptosis |
| Chromobox homolog 4 | -0.55 | Chromatin assembly or disassembly/regulation of transcription, DNA-dependent |
| Chromodomain helicase DNA binding protein 4 | -0.69 | Chromatin assembly or disassembly/ regulation of transcription from Pol II promoter |
| Coatomer protein complex, subunit α | -0.53 | Intracellular protein transport/ER-to-Golgi transport |
| Coiled-coil domain containing 6 | -0.41 | Cell growth and/or maintenance |
| Cytochrome P450, family 2, subfamily B, polypeptide 6 | 0.40 | Electron transport |
| Cytokine-like nuclear factor n-pac | -0.58 | Pentose-phosphate shunt |
| DEAD box polypeptide 21 | -0.76 | RNA binding/ATP-dependent RNA helicase activity |
| DEAH box polypeptide 9 | -0.69 | Double-stranded RNA binding/ATP-dependent RNA helicase activity |
| Dedicator of cytokinesis 9 | -0.71 | DNA replication |
| -0.60 | ||
| Dentatorubral-pallidoluysian atrophy | -0.70 | Central nervous system development |
| -0.68 | ||
| Endothelial PAS domain protein 1 | -0.81 | Angiogenesis |
| Eukaryotic translation initiation factor 4γ, 1 | -0.41 | Protein biosynthesis/regulation of translational initiation |
| Fatty acid desaturase 2 | -0.49 | Fatty acid biosynthesis/fatty acid desaturation |
| Fibroblast growth factor receptor 1 | -0.56 | MAPKKK cascade/fibroblast growth factor receptor signaling pathway |
| Filamin A, α | -0.50 | Cell surface receptor-linked signal transduction/actin cytoskeleton organization |
| -0.58 | ||
| H3 histone, family 3A | -0.80 | Nucleosome structure |
| H41 | -0.52 | Cell proliferation |
| Heterogeneous nuclear ribonucleoproteinU-like 1 | -0.59 | RNA processing |
| Histone 1, H2bg | 0.51 | Nucleosome assembly |
| Insulin-like growth factor 2 | -1.27 | Regulation of cell cycle/cell proliferation/insulin receptor signaling pathway |
| -0.98 | ||
| Integrin, β4 | -0.70 | Cell-matrix adhesion/integrin-mediated signaling pathway |
| -0.81 | ||
| Integrin, β5 | -1.54 | Integrin-mediated cell adhesion |
| Interleukin 6 signal transducer | -0.91 | Cell surface receptor-linked signal transduction |
| -0.56 | ||
| -0.69 | ||
| IQ motif containing GTPase activating protein 1 | -0.67 | Assembly scaffold for complex involved in reorganization of actin cytoskeleton |
| Jumonji domain containing 2B | -0.44 | DNA binding, regulation of transcription |
| Lanosterol synthase | -0.52 | Steroid biosynthesis |
| Low-density lipoprotein receptor-related protein 5 | -0.50 | Signal transduction/positive regulation of cell proliferation |
| Lutheran blood group | -0.68 | Cell adhesion/signal transduction |
| Met proto-oncogene | -0.79 | Cell surface receptor-linked signal transduction/cell proliferation |
| MYC-associated zinc finger protein | -0.95 | Regulation of transcription, DNA-dependent |
| NAD kinase | -0.45 | Metabolism/phosphorylation |
| Neuropilin 2 | -0.51 | Vascular endothelial growth factor receptor activity |
| Nonfunctional folate binding protein | -0.88 | Binding protein |
| Nuclear mitotic apparatus protein 1 | -0.76 | Mitotic anaphase |
| Nuclear pore complex interacting protein | -0.42 | Nuclear pore complex |
| -0.56 | ||
| -0.56 | ||
| -0.42 | ||
| Nuclear receptor coactivator 2 | -0.75 | Regulation of transcription, DNA-dependent |
| Nuclear receptor coactivator 3 | -0.41 | Regulation of transcription, DNA-dependent |
| Nuclear receptor corepressor 2 | -0.81 | Regulation of transcription, DNA-dependent |
| Nuclear receptor subfamily 2, group F, member 6 | -0.79 | Regulation of transcription, DNA-dependent |
| Paxillin | -1.02 | Cell-matrix adhesion/signal transduction |
| Phosphatidic phosphatase type 2A | -0.40 | Dephosphorylation |
| Phosphatidylinositol-binding clathrin assembly protein | -0.59 | Protein complex assembly |
| Phosphodiesterase 4D, cAMP-specific | -0.69 | Cyclic nucleotide metabolism |
| -0.56 | ||
| -0.84 | ||
| Plakophilin 2 | -0.94 | Cell-cell adhesion |
| Plectin 1, intermediate filament binding protein, 500 kDa | -0.77 | Cytoskeletal anchoring |
| Poliovirus receptor-related 1 | -1.24 | Cell-cell adhesion |
| Proteasome activator subunit 4 | -0.51 | DNA repair |
| Protein tyrosine phosphatase, receptor type, O | -0.44 | Protein amino acid dephosphorylation |
| PTK9 protein tyrosine kinase 9 | -0.44 | Actin binding/phosphorylation |
| Rap guanine nucleotide exchange factor 1 | -0.70 | Small GTPase-mediated signal transduction |
| Rho GDP dissociation inhibitor α | -0.80 | Negative regulation of cell adhesion |
| Ribosomal protein L37a | -1.21 | Ribosomal protein |
| Ribosome binding protein 1 homolog, 180 kDa | -0.75 | Protein targeting/protein transport |
| Septin 11 | -0.59 | Cell cycle |
| SH3-domain GRB2-like endophilin B2 | -0.58 | Protein binding |
| SHC transforming protein 1 | -0.45 | Activation of MAPK/positive regulation of cell proliferation and mitosis |
| Small nuclear ribonucleoprotein polypeptide E | -0.69 | Component of spliceosome complex |
| Small optic lobes homolog | -0.51 | Proteolysis and peptidolysis |
| Solute carrier family 6, member 6 | -0.43 | Amino acid metabolism |
| Solute carrier family 7, member 2 | -0.88 | Amino acid metabolism |
| SON DNA binding protein | -0.57 | Anti-apoptosis |
| Sp1 transcription factor | -0.44 | Regulation of transcription, DNA-dependent |
| S-phase kinase-associated protein 2 | -0.47 | G1/S transition of mitotic cell cycle/cell proliferation |
| Spindlin | -0.47 | Gametogenesis |
| Stearoyl-CoA desaturase | -0.57 | Fatty acid synthesis/fatty acid desaturation |
| -0.69 | ||
| Steroid sulfatase, arylsulfatase C, isozyme S | -0.47 | Steroid catabolism |
| Striatin, calmodulin binding protein 4 | -0.56 | Signal transduction |
| Tetratricopeptide repeat domain 3 | -0.58 | Protein ubiquitination |
| Thymopoietin | -0.88 | DNA binding |
| Translocated promoter region | -0.56 | Protein-nucleus import |
| Tropomodulin 3 | -0.54 | Actin and tropomyosin binding |
| UDP-Gal:βGlcNAc β 1,4-galactosyltransferase, polypeptide 1 | -0.74 | Carbohydrate metabolism |
| V-akt murine thymoma viral oncogene homolog 2 | -0.79 | Protein amino acid phosphorylation |
| -0.88 | ||
| WD repeat and SOCS box-containing 1 | -0.53 | Intracellular signaling cascade |
| WW domain containing adaptor with coiled-coil | -0.64 | Protein-protein interactions/signaling |
| XPA binding protein 2 | -0.73 | DNA repair/transcription-coupled nucleotide-excision repair |
| Zinc finger protein 36, C3H type-like 1 | -0.87 | Transcription factor activity |
Fig. 2.
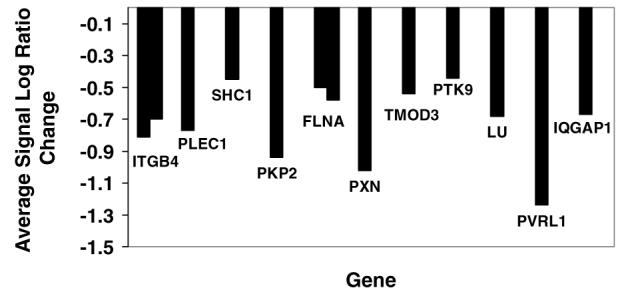
Histogram of repressed genes in microarray involved in cytoskeleton and cell-to-cell adhesion
RT-PCR analysis
To verify expression changes seen from microarray results, mRNA levels of selected genes, fatty acid desaturase 2 (FADS2), phospholipid hydroperoxidase (GPX4), insulin-like growth factor 2 (IGF2), gastrointestinal glutathione peroxidase (GPX2), glutathione reductase (GSR), and cytochrome P450, family 2, subfamily B, polypeptide 6 (CYP2B6), were analyzed using semi-quantitative RT-PCR. FADS2 and IGF2 were selected because they showed a decrease in expression. CYP2B6 was chosen because it showed an increase in expression. GPX4 was selected because it showed no change in expression. GPX2 was selected because Caco-2 cells mimic the gastrointestinal epithelium. GSR was selected because it showed no change in expression in one experiment but showed a decrease in expression in the other experiment. The band intensities of the PCR products for each gene were quantified for each cycle, the fold change for each gene was calculated for each cycle, each calculated fold change was normalized, and then the average fold change for each gene was found.
Data showed that fold changes detected in both microarray and RT-PCR were comparable. GPX4 and GPX2, which showed no change in expression in both microarray experiments, revealed changes of expression calculated to be ±10% in the RT-PCR experiments, indicating basically no change. Replicate data generated from the microarray and from the RT-PCR methods showed that CYP2B6 expression increased in cells treated with H2O2 compared to the control, although RT-PCR detected a higher percentage change. In the first GSR experiment, both microarray and RT-PCR data indicated a decrease in expression; however, the fold change in repression was greater in the microarray data compared to the RT-PCR data. Microarray and RT-PCR data for GSR in the second experiment similarly showed no change. Similarly for FADS2 and IGF2, both microarray and RT-PCR data in both experiments indicated a decrease in expression in the H2O2 treatment group compared to the control; however, the fold change in repression was larger in magnitude in the microarray data compared to the RT-PCR data.
Discussion
Methodology interpretation
In this study, the first to apply oligonucleotide microarray techniques for evaluating the effects of H2O2 treatment on the transcriptome of Caco-2 cells, expression levels in both microarray experiments of several genes of interest were confirmed by highly sensitive and specific semi-quantitative RT-PCR [17]. Quantification took place during the exponential phase and not the plateau phase [17]. Although the absolute values of fold induction/repression were not identical to those in the arrays, similar trends were observed. For example, comparable results between the arrays and RT-PCRs have been reported [5, 12, 17].
For this study, the criterion for change in gene expression was lowered to a signal log ratio of ±0.40. Other scientists have noted [9, 10] that genes with smaller differences in expression ratios are reliable and should be considered for analysis, especially when replicate experiments are performed and confirmed by RT-PCR. Genes that show only a small change in expression levels may be the most relevant for product formation, resulting in enormous changes in metabolism. Perhaps each gene has a certain rate equation for the equilibrium maintained between mRNA levels and the corresponding enzyme or protein. There is also a high degree of variability in translational efficiency among the different mRNAs [9]. Thus, a lower signal log ratio as the criterion for change in expression can be used as long as the change in expression is verified by RT-PCR or other appropriate methods.
Because the goal of this study was to identify genes that were altered across a population, RNA samples were pooled to reduce expression changes introduced by individual natural Caco-2 sample variability as reported in other experiments using human tissue, including duodenal, composed of different cell types [5, 16] and using cell culture samples from different or the same passage levels [16], including Caco-2 cells. It has also been found that fold ratios calculated from pooled comparisons correlated better with genes with significant p-values than fold ratios obtained from average comparisons [16]. Expression changes in cell culture are also influenced by cell density, cell viability, stage of cell cycle, and stage of cell growth [7]. Standardization of a test procedure, as in this study, is therefore necessary in order to identify consistent changes between replicate experiments.
Physiology interpretation
The current study exemplified the use of microarrays for the identification of a differential gene expression profile in Caco-2 cells in response to H2O2 treatment. These results provide evidence of the variety of genes that are affected by oxidation. When the genes showing altered expression were categorized according to their known function, it appeared that H2O2 down-regulated the expression of several genes that possibly could be involved in cell proliferation and growth. Inhibition of lanosterol synthase (LSS) and fatty acid desaturase 2 (FADS2) and of stearoyl-CoA desaturase (SCD) results in a decrease in endogenous cholesterol and unsaturated fatty acids, respectively. Since these lipids are components of cell membranes and cell proliferation involves the formation of cell membranes, the cell is unable to divide if these necessary components are not available [18-20].
Evidence shows that repression of genes involved in cell cycle and cell growth has detrimental effects on cell proliferation success. S-phase kinase-associated protein 2 (SKP2) is an essential element of cyclin A-CDK2 S-phase kinase, which is required for the G1-to-S phase transition in the mitotic cell cycle [21]. Mariadason et al. demonstrated that a decrease in Caco-2 cell proliferation correlates with a lower percentage of these cells in the S phase [9]. Insulin-like growth factor 2 (IGF2) is a potent mitogen for in vitro cultured cells that promotes cell growth and proliferation. It is known as a progression factor and is required after the competence factors to promote cell proliferation [22]. In addition, integrin β4 possesses oncogenic and growth-promoting activity [23]. Nuclear mitotic apparatus protein 1 (NUMA1) is involved in reassembling the nucleus in late mitosis, during anaphase, and after the chromosomes have been captured, aligned, and separated [24]. Dedicator of cytokinesis 9 (DOCK9) is involved in DNA replication and is a component of the kinesin complex required for cytokinesis [25]. Cell proliferation cannot proceed if the cell cannot enter S phase and replicate DNA and if the cell cannot ultimately divide.
It has been shown that proliferating cells, including tumor cells, are able to induce angiogenesis required for growth [9]. Vascular endothelial growth factor (VEGF) and the VEGF receptor, neuropilin, both play important roles in angiogenesis and cell growth [11]. The repression of endothelial PAS domain protein 1 (EPAS1), a transcription protein that regulates and enhances VEGF expression and thus vascularization [26], and neuropilin 2 (NRP2), a high-affinity VEGF receptor [27], indicates an inhibition of cell growth and proliferation.
As cell proliferation ceases, various components of basal transcription could be down-regulated as was found in our research. MYC-associated zinc finger protein (MAZ), a DNA specific transcription factor, is a potent regulator of cell cycle progression and cell proliferation, and according to our studies, its down-regulation is critical for growth inhibition [9]. Also, zinc finger transcription factor Sp1 (SP1) binds to GC box promoter elements and selectively activates mRNA synthesis [28]. Other repressed genes involved in the regulation of transcription were nuclear receptor co-activator 2 (NCOA2) and nuclear receptor coactivator 3 (NCOA3), which are both involved in transcription coactivation [29]. The latter is involved in histone acetyltransferase activity, which promotes histone acetylation and dissociation from DNA. NCOA3 permits transcription by the recruitment of p300/CBP-associated factor and CREB binding protein as part of a multisubunit coactivation complex, and is involved in the coactivation of the NF-κB pathway by interacting with the NF-κB1 subunit [29]. Repression of these genes suggests a decrease in transcription. As cell proliferation decreases, genes involved in DNA repair are down-regulated [9], as was shown in this study by the repression of XPA binding protein 2 (XAB2) and proteasome activator subunit 4 (PSME4). XAB2 is associated with RNA polymerase II and transcription-coupled nucleotide-excision repair [30]. PSME4 is involved in DNA repair possibly by recruiting proteasomes to double strand breaks [31].
Down-regulation of components involved in RNA processing and translation is also a consequence of cell proliferation cessation [9]. DEAD box polypeptide 21 (DDX21) [32] and DEAH box polypeptide 9 (DHX9) [33] are RNA helicases. Both polypeptides unwind and fold RNA to generate multiple RNA secondary structures that then influence RNA-binding proteins [9]. In addition, DDX21 and DHX9 could be involved in RNA editing and splicing in the nucleus. Small nuclear ribonucleoprotein (sn-RNP) polypeptide E (SNRPE) is part of the spliceosome complex. It associates with the sn-RNPs U1, U2, U4-U6, and U5, which are all implicated in splicing [34]. After RNA editing, the RNA must be transported to the cytoplasm. A nuclear RNA-binding protein of the heterogeneous nuclear ribonucleoprotein (hnRNP) family that may have a role in nucleocytoplasmic RNA transport is hnRNP U-like 1 (HNRPUL1), which was down-regulated in this experiment [9]. Also, the translocated promoter region (TPR) gene has many roles in the intranuclear dynamics of RNA polymerase II transcripts, including their processing, intranuclear transport, and translocation through nuclear pore complexes to the cytoplasm [35]. Gene expression of nuclear pore complex interacting proteins (NPIP) [36], ribosomal protein L37a (RPL37A), a component of the large 60S subunit [37], and ribosome binding protein 1 (RRBP1), a ribosome receptor-like protein that mediates the interaction between the ribosome and the endoplasmic reticulum membrane [38] was repressed.
One gene involved in the mediation of translation initiation, eukaryotic translation initiation factor 4γ, 1 (EIF4G1), was down-regulated. EIF4G1 is an interface or scaffolding protein that binds the poly(A)-binding protein component of the initiation factors eIF4A, eIF4E, and eIF3 and has a binding site for the mRNA cap [39]. This protein is involved in recruiting mRNA to the ribosome. Solute carrier family 7, member 2 (SLC7A2) is a low-affinity, high-capacity permease involved in the transport of the cationic or basic amino acids after their metabolism [5]. Its gene expression was down-regulated along with aspartyl-tRNA synthetase (DARS). A decrease in any synthetase can initiate a constricted response of an increase in uncharged tRNAs and blocks further progress by the ribosome, thus decreasing the efficiency of translation.
After synthesis, proteins must be transported throughout the cell to specific organelles. ATPase, H+ transporting, lysosomal (ATP6VOE), and coatomer protein complex, subunit α (COPA) both decreased in expression after the H2O2 treatment. The former protein is a component of vacuolar ATPase and is important for sorting proteins [40]. The latter protein is a part of a non-clathrin-coated, vesicular-coated complex and is involved in the transport of proteins between the endoplasmic reticulum and the Golgi apparatus [41]. Coatomer protein complex can only be recruited to membranes associated with ADP-ribosylation factors, which are small GTP-binding proteins. ADP-ribosylation factor 3 (ARF3) is associated with intracellular protein transport from the Golgi apparatus [42]. Its expression was also down-regulated by H2O2.
Phosphodiesterase 4D, cAMP-specific (PDE4D) [43] is the enzyme catalyzing the breakdown of cAMP to AMP. Down-regulation of PDE4D allows cAMP, a second messenger, to accumulate in the cell and then cAMP-dependent protein kinase is activated. This kinase then activates phosphorylase kinase, resulting in the phosphorylation of glycogen phosphorylase. This phosphorylation activates the enzyme, therefore causing the breakdown of glycogen into glucose [43]. Glucose provides, in this situation, the much needed cellular energy when the cell is under stress from oxidation.
During cell division and cell proliferation proteins associated with the actin cytoskeleton, the cell membrane skeleton, and cell-to-cell adhesion are needed. Down-regulation of genes representing integrin β4, SHC transforming protein 1 (SHC1), paxillin (PXN), plakophilin 2 (PKP2), Lutheran blood group (LU), poliovirus receptor-related 1 (PVRL1), plectin 1 (PLEC1), tropomodulin 3 (TMOD3), protein tyrosine kinase 9 (PTK9), filamin A, α (FLNA), and IQ motif containing GTPase activating protein 1 (IQGAP1) could indicate an inhibition of cell growth. ITGB4 is involved in the formation of hemidesmosomes of epithelial cells and forms very stable integrin-mediated cell adhesions to the extracellular matrix [44]. ITGB4 mediates transduced signals that regulate cell growth. SHC1 is a ras adapter that is associated with a particular phosphorylation tyrosine pattern on ITGB4 [44]. By attaching to ITGB4, a receptor tyrosine kinase, SHC1 is a positive regulator of cell proliferation and cell mitosis or division [45]. ITGB4 is also linked to the structural protein PXN that is localized to focal adhesions, which are the sites of integrin-mediated cell adhesion to the extracellular matrix [46]. PKP2 is a structural protein associated with desmosomes and cell-to-cell adhesion [47]. PLEC1 is an intracellular component of the hemidesmosome involved in cytoskeletal anchoring. It is an intermediate filament binding protein that cross-links intermediate filaments and connects them to other cell structures and other cells [48]. TMOD3 is a component of the junctional complex in the cytoskeleton of the cell membrane binding with actin and tropomyosin [49]. PTK9 [50] and FLNA are cross-linking actin binding proteins that make up the actin cytoskeleton. FLNA links actin filaments to membrane glycoproteins, serving as a scaffold for a wide range of cytoplasmic signaling proteins, such as NF-κB [51]. IQGAP1 [52] associates with calmodulin and serves as an assembly scaffold for the organization of a multimolecular complex. This complex coordinates incoming signals to the reorganization of the actin cytoskeleton at the plasma membrane. No reorganization of the actin cytoskeleton at the plasma membrane is needed if cells do not proliferate as was in this case.
UDP-Gal:βGlcNAc β1,4-galactosyltransferase, polypeptide 1 (B4GALT1) is responsible for synthesizing certain cell surface glycolipids and glycoproteins that function as recognition molecules for cell to cell and cell to matrix interactions [53]. Oxidation of Caco-2 cells by H2O2 down-regulated B4GALT1 expression as well, implicating a discontinuance of cell-to-cell communication and cell growth.
Receptor tyrosine kinases (RTKs) play a critical role in the stimulation of proliferation by binding growth factors. After the dimerization of RTKs, their autophosphorylation begins an intracellular signaling cascade that instigates or initiates cell proliferation and growth. ITGB4 [23], met proto-oncogene (MET) [54], interleukin 6 signal transducer (IL6ST) [55], fibroblast growth factor receptor 1 (FGFR1) [56], and low-density lipoprotein receptor-related protein 5 (LRP5) [57] are cell surface receptors—and all but the last are RTK-like—that are positive regulators of cell proliferation. The gene expression of these receptors was down-regulated by H2O2 in this experiment, indicating a halt in cell proliferation. Following autophosphorylation, RTKs transmit the intracellular signal via proteins containing Src homology 2 (SH2) and 3 (SH3) domains. Consistent with the down-regulation of several RTKs, both SH2-domain SHC1 and SH3-domain GRB2-like endophilin B2 (SH3GLB2) [58] mRNAs were down-regulated as well. A guanine nucleotide exchange factor (GEF) then induces Ras to exchange its bound GDP for GTP, thus activating it. Again, in this experiment, Rap GEF (RAPGEF1) [59], which binds to the SH3 domain of GRB2/ASH and transduces the signal to activate Ras, was down-regulated, indicative of cessation of cell proliferation. Direct interaction between Ras-GTP and a serine/threonine kinase then transduces the signal downstream, which consists of a linear cascade of serine/threonine kinases ending in the phosphorylation of nuclear transcription factors by a mitogen-activated protein kinase (MAPK). One rac-B-serine/threonine kinase repressed by H2O2 was v-akt murine thymoma viral oncogene homolog 2 (AKT2) [60]. Ultimately, the nuclear transcription factor MYC-associated zinc finger protein was repressed.
Results of this microarray study provide the first global assessment of H2O2-sensitive genes in Caco-2 cells. The decrease in expression of genes involved in lipid synthesis, cell cycle progression and cell division, angiogenesis, transcription and DNA repair, RNA processing and translation, protein transport and proteolysis, cAMP metabolism, cytoskeleton and cell-to-cell adhesion, RTKs, and intracellular and extracellular signaling may provide insight into the molecular mechanisms underlying the inhibition of cell growth and proliferation in Caco-2 cells after oxidation with H2O2.
Acknowledgments
This study is published as Journal Series No. 15123, Nebraska Agricultural Research Division, Department of Food Science and Technology, University of Nebraska—Lincoln. The work was supported by the Agricultural Research Division of the University of Nebraska—Lincoln and NIH Grants DK 60447 and DK 063945.
Contributor Information
Theresa A. Herring, Department of Nutrition and Health Sciences, University of Nebraska—Lincoln, Lincoln, Nebraska 68583, USA
Susan L. Cuppett, Department of Food Science and Technology, University of Nebraska—Lincoln, Lincoln, Nebraska 68583, USA
Janos Zempleni, Department of Nutrition and Health Sciences, University of Nebraska—Lincoln, Lincoln, Nebraska 68583, USA.
References
- 1.Engler JA, Gupta A, Li L, Rao RK. Inhibition of DNA synthesis in Caco-2 cells by oxidative stress. Amelioration by epidermal growth factor. Dig Dis Sci. 1999;44:1902–1909. doi: 10.1023/a:1018815327769. [DOI] [PubMed] [Google Scholar]
- 2.Bestwick CS, Milne L. Alteration of culture regime modifies antioxidant defenses independent of intracellular reactive oxygen levels and resistance to severe oxidative stress within confluent Caco-2 “intestinal cells. Dig Dis Sci. 2001;46:417–423. doi: 10.1023/a:1005621403626. [DOI] [PubMed] [Google Scholar]
- 3.Tadjali M, Seidelin JB, Olsen J, Troelsen JT. Transcriptome changes during intestinal cell differentiation. Biochim Biophys Acta. 2002;1589:160–167. doi: 10.1016/s0167-4889(02)00170-2. [DOI] [PubMed] [Google Scholar]
- 4.Peters WHN, Roelofs HMJ. Time-dependent activity and expression of glutathione S-transferases in the human colon adenocarcinoma cell line Caco-2. Biochem J. 1989;264:613–616. doi: 10.1042/bj2640613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun D, Lennernas H, Welage LS, Barnett JL, Landowski CP, Foster D, Fleisher D, Lee K, Amidon GL. Comparison of human duodenum and Caco-2 gene expression profiles for 12,000 gene sequence tags and correlation with permeability of 26 drugs. Pharm Res. 2002;19:1400–1416. doi: 10.1023/a:1020483911355. [DOI] [PubMed] [Google Scholar]
- 6.Desikan R, Mackerness SAH, Hancock JT, Neill SJ. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001;127:159–172. doi: 10.1104/pp.127.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aherne SA, O’Brien NM. Protection by the flavonoids myricetin, quercetin, and rutin against hydrogen peroxide-induced DNA damage in Caco-2 and Hep G2 cells. Nutr Cancer. 1999;34:160–166. doi: 10.1207/S15327914NC3402_6. [DOI] [PubMed] [Google Scholar]
- 8.Rao RK, Li L, Baker RD, Baker SS, Gupta A. Glutathione oxidation and PTPase inhibition by hydrogen peroxide in Caco-2 cell monolayer. Am J Physiol Gastrointest Liver Physiol. 2000;279:G332–G340. doi: 10.1152/ajpgi.2000.279.2.G332. [DOI] [PubMed] [Google Scholar]
- 9.Mariadason JM, Arango D, Corner GA, Aranes MJ, Hotchkiss KA, Yang W, Augenlicht LH. A gene expression profile that defines colon cell maturation in vitro. Cancer Res. 2002;62:4791–4804. [PubMed] [Google Scholar]
- 10.Blanchard RK, Moore JB, Green CL, Cousins RJ. Modulation of intestinal gene expression by dietary zinc status: effectiveness of cDNA arrays for expression profiling of a single nutrient deficiency. Proc Natl Acad Sci USA. 2001;98:13507–13513. doi: 10.1073/pnas.251532498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Sarkar FH. Gene expression profiles of genistein-treated PC3 prostate cancer cells. J Nutr. 2002;132:3623–3631. doi: 10.1093/jn/132.12.3623. [DOI] [PubMed] [Google Scholar]
- 12.Roy S, Lado BH, Khanna S, Sen CK. Vitamin E sensitive genes in the developing rat fetal brain: a high-density oligonucleotide microarray analysis. FEBS Lett. 2002;530:17–23. doi: 10.1016/s0014-5793(02)03309-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton JL, Madsen SA, Yao J, Sipkovsky SS, Coussens PM. An immunogenomics approach to understanding periparturient immunosuppression and mastitis susceptibility in dairy cows. Acta Vet Scand. 2001;42:407–424. [PubMed] [Google Scholar]
- 14.Kim J, Kang YS, Lee SH, Lee E, Yoo BH, Lee YS. Gliben-clamide induces apoptosis through inhibition of cystic fibrosis transmembrane conductance regulator (CFTR) Cl- channels and intracellular Ca2+ release in HepG2 human hepatoblastoma cells. Biochem Biophys Res Commun. 1999;261:682–688. doi: 10.1006/bbrc.1999.1108. [DOI] [PubMed] [Google Scholar]
- 15.Hirschi KD, Kreps JA, Hirschi KK. Molecular approaches to studying nutrient metabolism and function: an array of possibilities. J Nutr. 2001;131:1605S–1609S. doi: 10.1093/jn/131.5.1605S. [DOI] [PubMed] [Google Scholar]
- 16.Kaminski N, Friedman N. Practical approaches to analyzing results of microarray experiments. Am J Respir Cell Mol Biol. 2002;27:125–132. doi: 10.1165/ajrcmb.27.2.f247. [DOI] [PubMed] [Google Scholar]
- 17.Pfaffl MW, Gerstmayer B, Bosio A, Windisch W. Effect of zinc deficiency on the mRNA expression pattern in liver and jejunum of adult rats: monitoring gene expression using cDNA microarrays combined with real-time RT-PCR. J Nutr Biochem. 2003;14:691–702. doi: 10.1016/j.jnutbio.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Corey EJ, Matsuda SPT, Bartel B. Molecular cloning, characterization, and overexpression of ERG7, the Saccharomyces cerevisiae gene encoding lanosterol synthase. Proc Natl Acad Sci USA. 1994;91:2211–2215. doi: 10.1073/pnas.91.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho HP, Nakamura MT, Clarke SD. Cloning, expression, and nutritional regulation of the mammalian Δ-6 desaturase. J Biol Chem. 1999;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- 20.Scaglia N, Igal RA. Stearoyl-CoA desaturase is involved in the control of proliferation, anchorage-independent growth, and survival in human transformed cells. J Biol Chem. 2005;280:25339–25349. doi: 10.1074/jbc.M501159200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]
- 22.Bell GI, Merryweather JP, Sanchez-Pescador R, Stempien MM, Priestley L, Scott J, Rall LB. Sequence of a cDNA clone encoding human preproinsulin-like growth factor II. Nature. 1984;310:775–777. doi: 10.1038/310775a0. [DOI] [PubMed] [Google Scholar]
- 23.Russell AJ, Fincher EF, Millman L, Smith R, Vela V, Waterman EA, Dey CN, Guide S, Weaver VM, Marinkovich MP. Alpha 6 beta 4 integrin regulates keratinocyte chemotaxis through differential GTPase activation and antagonism of alpha 3 beta 1 integrin. J Cell Sci. 2003;116:3543–3556. doi: 10.1242/jcs.00663. [DOI] [PubMed] [Google Scholar]
- 24.Lydersen BK, Pettijohn DE. Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell. 1980;22:489–499. doi: 10.1016/0092-8674(80)90359-1. [DOI] [PubMed] [Google Scholar]
- 25.Meller N, Irani-Tehrani M, Kiosses WB, Del Pozo MA, Schwartz MA. Zizimin1, a novel Cdc42 activator, reveals a new GEF domain for Rho proteins. Nat Cell Biol. 2002;4:639–647. doi: 10.1038/ncb835. [DOI] [PubMed] [Google Scholar]
- 26.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 27.Takashima S, Kitakaze M, Asakura M, Asanuma H, Sanada S, Tashiro F, Niwa H, Miyazaki Ji J, Hirota S, Kitamura Y, Kitsukawa T, Fujisawa H, Klagsbrun M, Hori M. Targeting of both mouse neuropilin-1 and neuropilin-2 genes severely impairs developmental yolk sac and embryonic angiogenesis. Proc Natl Acad Sci USA. 2002;99:3657–3662. doi: 10.1073/pnas.022017899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cadoret A, Baron-Delage S, Bertrand F, Kornprost M, Groyer A, Gespach C, Capeau J, Cherqui G. Oncogene-induced up-regulation of Caco-2 cell proliferation involves IGF-II gene activation through a protein kinase C-mediated pathway. Oncogene. 1998;17:877–887. doi: 10.1038/sj.onc.1202013. [DOI] [PubMed] [Google Scholar]
- 29.Zhou G, Hashimoto Y, Kwak I, Tsai SY, Tsai MJ. Role of the steroid receptor coactivator SRC-3 in cell growth. Mol Cell Biol. 2003;23:7742–7755. doi: 10.1128/MCB.23.21.7742-7755.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakatsu Y, Asahina H, Citterio E, Rademakers S, Vermeulen W, Kamiuchi S, Yeo JP, Khaw MC, Saijo M, Kodo N, Matsuda T, Hoeijmakers JH, Tanaka K. XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J Biol Chem. 2000;275:34931–34937. doi: 10.1074/jbc.M004936200. [DOI] [PubMed] [Google Scholar]
- 31.Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21:3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valdez BC, Henning D, Busch RK, Woods K, Flores-Rozas H, Hurwitz J, Perlaky L, Busch H. A nucleolar RNA helicase recognized by autoimmune antibodies from a patient with watermelon stomach disease. Nucleic Acids Res. 1996;24:1220–1224. doi: 10.1093/nar/24.7.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tetsuka T, Uranishi H, Sanda T, Asamitsu K, Yang JP, Wong-Staal F, Okamoto T. RNA helicase A interacts with nuclear factor kappaB p65 and functions as a transcriptional coactivator. Eur J Biochem. 2004;271:3741–3751. doi: 10.1111/j.1432-1033.2004.04314.x. [DOI] [PubMed] [Google Scholar]
- 34.Stanford DR, Kehl M, Perry CA, Holicky EL, Harvey SE, Rohleder AM, Rehder K, Jr, Luhrmann R, Wieben ED. The complete primary structure of the human snRNP E protein. Nucleic Acids Res. 1988;16:10593–10605. doi: 10.1093/nar/16.22.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krull S, Thyberg J, Bjorkroth B, Rackwitz HR, Cordes VC. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell. 2004;15:4261–4277. doi: 10.1091/mbc.E04-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 37.Saha DP, Tirumalai PS, Scala LA, Howells RD. Human ribosomal protein L37a: a cloning of the cDNA and analysis of differential gene expression in tissues and cell lines. Gene. 1993;132:285–289. doi: 10.1016/0378-1119(93)90209-l. [DOI] [PubMed] [Google Scholar]
- 38.Diefenbach RJ, Diefenbach E, Douglas MW, Cunningham AL. The ribosome receptor, p180, interacts with kinesin heavy chain, KIF5B. Biochem Biophys Res Commun. 2004;319:987–992. doi: 10.1016/j.bbrc.2004.05.069. [DOI] [PubMed] [Google Scholar]
- 39.Gradi A, Imataka H, Svitkin YV, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simckes AM, Swanson SK, White RA. Chromosomal localization of three vacuolar-H+-ATPase 16 kDa subunit (ATP6VOC) genes in the murine genome. Cytogenet Genome Res. 2002;97:111–115. doi: 10.1159/000064065. [DOI] [PubMed] [Google Scholar]
- 41.Presley JF, Ward TH, Pfeifer AC, Siggia ED, Phair RD, Lippincott-Schwartz J. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- 42.Takatsu H, Yoshino K, Toda K, Nakayama K. GGA proteins associate with Golgi membranes through interaction between their GGAH domains and ADP-ribosylation factors. Biochem J. 2002;365:369–378. doi: 10.1042/BJ20020428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landells LJ, Szilagy CM, Jones NA, Banner KH, Allen JM, Doherty A, O’Connor BJ, Spina D, Page CP. Identification and quantification of phosphodiesterase 4 subtypes in CD4 and CD8 lymphocytes from healthy and asthmatic subjects. Br J Pharmacol. 2001;133:722–729. doi: 10.1038/sj.bjp.0704120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frantz DJ, Hughes BG, Nelson DR, Murray BK, Christensen MJ. Cell cycle arrest and differential gene expression in HT-29 cells exposed to an aqueous garlic extract. Nutr Cancer. 2000;38:255–264. doi: 10.1207/S15327914NC382_15. [DOI] [PubMed] [Google Scholar]
- 45.Pelicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Grignani F, Pawson T, Pelicci PG. A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]
- 46.Salgia R, Li JL, Lo SH, Brunkhorst B, Kansas GS, Sobhany ES, Sun Y, Pisick E, Hallek M, Ernst T, Tantravahi R, Chen LB, Griffin JD. Molecular cloning of human paxillin, a focal adhesion protein phosphorylated by P210BCR/ABL. J Biol Chem. 1995;270:5039–5047. doi: 10.1074/jbc.270.10.5039. [DOI] [PubMed] [Google Scholar]
- 47.Grossmann KS, Grund C, Huelsken J, Behrend M, Erdmann B, Franke WW, Birchmeier W. Requirement of plakophilin 2 for heart morphogenesis and cardiac junction formation. J Cell Biol. 2004;167:149–160. doi: 10.1083/jcb.200402096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geerts D, Fontao L, Nievers MG, Schaapveld RQ, Purkis PE, Wheeler GN, Lane EB, Leigh IM, Sonnenberg A. Binding of integrin alpha6beta4 to plectin prevents plectin association with F-actin but does not interfere with intermediate filament binding. J Cell Biol. 1999;147:417–434. doi: 10.1083/jcb.147.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee A, Fischer RS, Fowler VM. Stabilization and remodeling of the membrane skeleton during lens fiber cell differentiation and maturation. Dev Dyn. 2000;217:257–270. doi: 10.1002/(SICI)1097-0177(200003)217:3<257::AID-DVDY4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 50.Beeler JF, LaRochelle WJ, Chedid M, Tronick SR, Aaronson SA. Prokaryotic expression cloning of a novel human tyrosine kinase. Mol Cell Biol. 1994;14:982–988. doi: 10.1128/mcb.14.2.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang M, Breitwieser GE. High affinity interaction with filamin A protects against calcium-sensing receptor degradation. J Biol Chem. 2005;280:11140–11146. doi: 10.1074/jbc.M412242200. [DOI] [PubMed] [Google Scholar]
- 52.Swart-Mataraza JM, Li Z, Sacks DB. IQGAP1 is a component of Cdc42 signaling to the cytoskeleton. J Biol Chem. 2002;277:24753–24763. doi: 10.1074/jbc.M111165200. [DOI] [PubMed] [Google Scholar]
- 53.Kotani N, Asano M, Iwakura Y, Takasaki S. Knockout of mouse beta 1,4-galactosyltransferase-1 gene results in a dramatic shift of outer chain moieties of N-glycans from type 2 to type 1 chains in hepatic membrane and plasma glycoproteins. Biochem J. 2001;357:827–834. doi: 10.1042/0264-6021:3570827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coltella N, Manara MC, Cerisano V, Trusolino L, Di Renzo MF, Scotlandi K, Ferracini R. Role of the MET/HGF receptor in proliferation and invasive behavior of osteosarcoma. FASEB J. 2003;17:1162–1164. doi: 10.1096/fj.02-0576fje. [DOI] [PubMed] [Google Scholar]
- 55.Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 56.Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 57.Dong Y, Lathrop W, Weaver D, Qiu Q, Cini J, Bertolini D, Chen D. Molecular cloning and characterization of LR3, a novel LDL receptor family protein with mitogenic activity. Biochem Biophys Res Commun. 1998;251:784–790. doi: 10.1006/bbrc.1998.9545. [DOI] [PubMed] [Google Scholar]
- 58.Cuddeback SM, Yamaguchi H, Komatsu K, Miyashita T, Yamada M, Wu C, Singh S, Wang HG. Molecular cloning and characterization of Bif-1. A novel Src homology 3 domain-containing protein that associates with Bax. J Biol Chem. 2001;276:20559–20565. doi: 10.1074/jbc.M101527200. [DOI] [PubMed] [Google Scholar]
- 59.Hirata T, Nagai H, Koizumi K, Okino K, Harada A, Onda M, Nagahata T, Mikami I, Hirai K, Haraguchi S, Jin E, Kawanami O, Shimizu K, Emi M. Amplification, up-regulation and overexpression of C3G (CRK SH3 domain-binding guanine nucleotide-releasing factor) in non-small cell lung cancers. J Hum Genet. 2004;49:290–295. doi: 10.1007/s10038-004-0148-1. [DOI] [PubMed] [Google Scholar]
- 60.Cheng JQ, Godwin AK, Bellacosa A, Taguchi T, Franke TF, Hamilton TC, Tsichlis PN, Testa JR. AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas. Proc Natl Acad Sci USA. 1992;89:9267–9271. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]