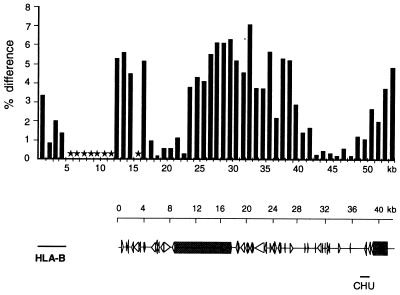
Figure 3.
Sequence comparison of allelic regions between HLA-B and -C. The sequence of cosmid Y5C028 was compared over the first 3 kb with cosmid O32A, which was derived from a non-CGM1 HLA haplotype (24) and over positions 4,000–42,000 with cosmid Y3C071, derived from YAC 3. cross_match analysis in sequential 1-kb blocks of Y5C028 against O32A or Y3C071 was used with the following parameters: minmatch 20, minscore 40, bandwidth 14, indexwordsize 10, masklevel 80. Y5C028 and Y3C071 span the region between HLA-B and -C identified by MCD mapping as highly variable between the two CGM1 chromosomes (shaded region in Fig. 2). Each bar represents the percentage difference over the 1-kb region compared. Included at the left of the bar graph is a similar comparison of the divergence between the HLA-B locus contained in O32A and a genomic HLA-B locus from the database. Below is a depiction of the genetic content including genome wide repeats and other homologies as discussed in the text, aligned to scale with the bar graph. Solid arrows indicate alu repeats, open arrows indicate MIR repeats, and shaded boxes indicate reverse transcriptase homologies. CHU, class I homology unit.