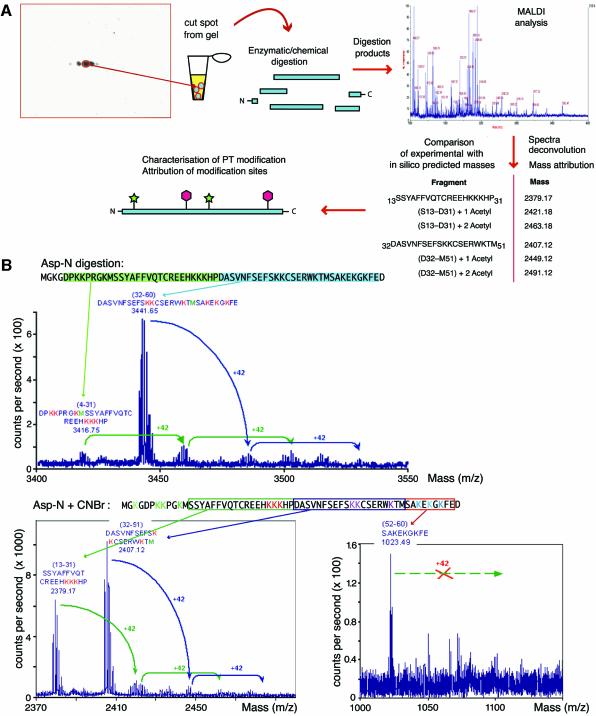
Fig. 2. Strategy for 2D/MALDI-MS analysis of multiply modified HMGB1. (A) Spots were excised from 2D gels, proteolysed and analysed by MALDI-TOF. A mass was attributed to each peptide in the mixture; the masses corresponding to peptides predicted in silico were selected as ‘anchors’, and we searched the complex spectra for further peaks corresponding to anchor masses plus multiples of 42 (the molecular weight of an acetyl group). The analysis of two peptides is shown; the procedure was iterated for all peptides. We also mined the spectra for evidence of phosphorylation, methylation and glycosylation, without finding any. (B) An example of the multi-step digestion strategy developed to assign acetylation sites in HMGB1. Spots from 2D gels were digested in gel with protease Asp-N. One aliquot was analysed by MALDI-TOF: the arrows identify the peaks corresponding to unmodified fragment. We then looked for peaks with the molecular weight of unmodified fragments plus multiples of 42. The maximum number of acetyl moieties on each fragment was thus determined. Another aliquot of Asp-N digested HMGB1 was further digested with CNBr, and the products were analysed similarly. We proceeded with further cleavages until we could obtain identifiable fragments where all lysines were acetylated, or none was.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.