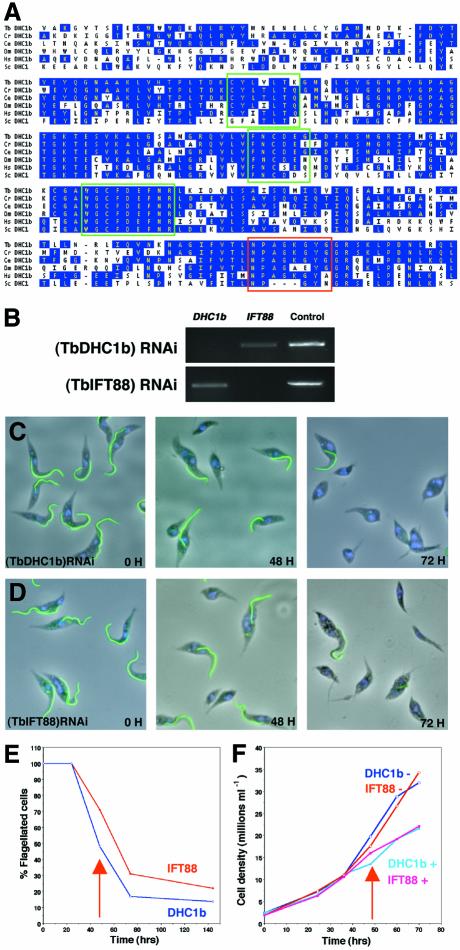
Fig. 1. TbDHC1b and TbIFT88 are required for flagellum assembly. (A) Alignment of protein sequences of the DHC1b central domain from T.brucei (Tb), Chlamydomonas reinhardtii (Cr), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (Hs) and the cytoplasmic dynein from Saccharomyces cerevisae (Sc). Conserved residues are shown in blue, and green and red blocks indicate dynein and DHC1b signatures, respectively. (B) RT–PCR analysis performed on total RNA extracted from non-induced and 48 h-induced (TbDHC1b) RNAi or (TbIFT88) RNAi mutants using specific primers for TbDHC1b, TbIFT88 or an unrelated RNA as a control. Knockdown of TbDHC1b (C) or of TbIFT88 (D) lead to production of non-flagellated cells over time. Cells were induced for the indicated periods and stained with the anti-PFRA antibody L8C4 as a marker of the flagellum (green), and with DAPI (blue) showing nucleus and kinetoplast. (E) RNAi was induced in (TbDHC1b) RNAi or (TbIFT88) RNAi cell lines, and cells were fixed at the indicated times and processed for immunofluorescence with anti-PFRA. Cells possessing at least one flagellum were scored as positive. Arrows indicate when non-flagellated cells were first detected. (F) In the same experiment, cell growth was monitored in the presence (+) or absence (–) of tetracycline.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.